A novel element upstream of the Vgamma2 gene in the murine T cell receptor gamma locus cooperates with the 3' enhancer to act as a locus control region
- PMID: 10477551
- PMCID: PMC2195622
- DOI: 10.1084/jem.190.5.669
A novel element upstream of the Vgamma2 gene in the murine T cell receptor gamma locus cooperates with the 3' enhancer to act as a locus control region
Abstract
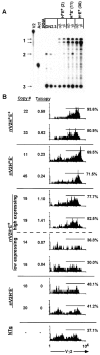
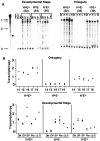
Transgenic expression constructs were employed to identify a cis-acting transcription element in the T cell receptor (TCR)-gamma locus, called HsA, between the Vgamma5 and Vgamma2 genes. In constructs lacking the previously defined enhancer (3'E(Cgamma1)), HsA supports transcription in mature but not immature T cells in a largely position-independent fashion. 3'E(Cgamma1), without HsA, supports transcription in immature and mature T cells but is subject to severe position effects. Together, the two elements support expression in immature and mature T cells in a copy number-dependent, position-independent fashion. Furthermore, HsA was necessary for consistent rearrangement of transgenic recombination substrates. These data suggest that HsA provides chromatin-opening activity and, together with 3'E(Cgamma1), constitutes a T cell-specific locus control region for the TCR-gamma locus.
Figures
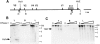

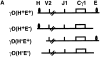

Similar articles
-
Redundant and unique roles of two enhancer elements in the TCRgamma locus in gene regulation and gammadelta T cell development.Immunity. 2002 Mar;16(3):453-63. doi: 10.1016/s1074-7613(02)00285-6. Immunity. 2002. PMID: 11911829
-
Interleukin 7 receptor control of T cell receptor gamma gene rearrangement: role of receptor-associated chains and locus accessibility.J Exp Med. 1998 Dec 21;188(12):2233-41. doi: 10.1084/jem.188.12.2233. J Exp Med. 1998. PMID: 9858510 Free PMC article.
-
Developmentally ordered V-J recombination in mouse T cell receptor gamma locus is not perturbed by targeted deletion of the Vgamma4 gene.J Immunol. 1997 May 1;158(9):4223-8. J Immunol. 1997. PMID: 9126983
-
Developmental regulation of V(D)J recombination at the TCR alpha/delta locus.Immunol Rev. 1998 Oct;165:131-47. doi: 10.1111/j.1600-065x.1998.tb01236.x. Immunol Rev. 1998. PMID: 9850858 Review.
-
[Gamma delta T cells--gene to biology].Tanpakushitsu Kakusan Koso. 1995 Oct;40(14):2118-25. Tanpakushitsu Kakusan Koso. 1995. PMID: 8532867 Review. Japanese. No abstract available.
Cited by
-
Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation.Blood. 2006 Jun 15;107(12):4898-906. doi: 10.1182/blood-2005-09-3596. Epub 2006 Feb 21. Blood. 2006. PMID: 16493008 Free PMC article.
-
Interleukin 15 controls the generation of the restricted T cell receptor repertoire of gamma delta intestinal intraepithelial lymphocytes.Nat Immunol. 2005 Dec;6(12):1263-71. doi: 10.1038/ni1267. Epub 2005 Nov 6. Nat Immunol. 2005. PMID: 16273100 Free PMC article.
-
The genomic arrangement of T cell receptor variable genes is a determinant of the developmental rearrangement pattern.Proc Natl Acad Sci U S A. 2004 Jan 6;101(1):260-5. doi: 10.1073/pnas.0303738101. Epub 2003 Dec 22. Proc Natl Acad Sci U S A. 2004. PMID: 14691262 Free PMC article.
-
Regulation of T-cell Receptor Gene Expression by Three-Dimensional Locus Conformation and Enhancer Function.Int J Mol Sci. 2020 Nov 11;21(22):8478. doi: 10.3390/ijms21228478. Int J Mol Sci. 2020. PMID: 33187197 Free PMC article. Review.
-
Defective development of gamma/delta T cells in interleukin 7 receptor-deficient mice is due to impaired expression of T cell receptor gamma genes.J Exp Med. 1999 Oct 4;190(7):973-82. doi: 10.1084/jem.190.7.973. J Exp Med. 1999. PMID: 10510087 Free PMC article.
References
-
- Sleckman B.P., Gorman J.R., Alt F.W. Accessibility control of antigen-receptor variable-region gene assemblyrole of cis-acting elements. Annu. Rev. Immunol. 1996;14:459–481. - PubMed
-
- Schatz D.G., Oettinger M.A., Schlissel M.S. V(D)J recombinationmolecular biology and regulation. Annu. Rev. Immunol. 1992;10:359–383. - PubMed
-
- Leiden J.M. Transcriptional regulation of T cell receptor genes. Annu. Rev. Immunol. 1993;11:539–570. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

