Insulin prohormone processing, distribution, and relation to metabolism in Aplysia californica
- PMID: 10479677
- PMCID: PMC6782465
- DOI: 10.1523/JNEUROSCI.19-18-07732.1999
Insulin prohormone processing, distribution, and relation to metabolism in Aplysia californica
Abstract
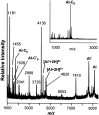
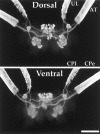
The first Aplysia californica insulin gene is characterized and its proteolytic processing from prohormone to final peptides elucidated using a combination of biochemical and mass spectrometric methods. Aplysia insulin (AI) is one of the largest insulins found, with a molecular weight of 9146 Da, and an extended A chain compared with other invertebrate and vertebrate insulins. The AI prohormone produces a series of C peptides and also a unique N-terminally acetylated D peptide. AI-producing cells are restricted to the central region of the cerebral ganglia mostly within the F and C clusters, and AI is transported to neurohemal release sites located on the upper labial and anterior tentacular nerves. The expression of AI mRNA decreases when the animal is deprived of food, and injections of AI reduce hemolymph glucose levels, suggesting that the function of insulin-regulating metabolism has been conserved.
Figures







References
-
- Abraham GN, Podell DN. Pyroglutamic acid: nonmetabolic formation, function in proteins and peptides, and characteristics of the enzymes effecting its removal. Mol Cell Biochem. 1981;38:181–190. - PubMed
-
- Banting FG, Best CH. The internal secretion of the pancreas. J Lab Clin Med. 1922;7:251–266. - PubMed
-
- Bateman A, Solomon S, Bennett HPJ. Post-translational modification of bovine pro-opiomelanocortin: tyrosine sulfation and pyroglutamate formation, a mass spectrometric study. J Biol Chem. 1990;266:22130–22136. - PubMed
-
- Bell GI, Pictet RL, Rutter WJ, Cordell B, Tischer E, Goodman HM. Sequence of the human insulin gene. Nature. 1960;284:26–32. - PubMed
-
- Boyd FT, Clarke DW, Mather TF, Raizada MK. Insulin receptors and insulin modulation of norepinephrine uptake in neuronal cultures from rat brain. J Biol Chem. 1985;260:15880–15884. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
