Holliday junction processing in bacteria: insights from the evolutionary conservation of RuvABC, RecG, and RusA
- PMID: 10482492
- PMCID: PMC94071
- DOI: 10.1128/JB.181.18.5543-5550.1999
Holliday junction processing in bacteria: insights from the evolutionary conservation of RuvABC, RecG, and RusA
Figures
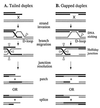
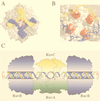



References
-
- Ariyoshi M, Vassylyev D G, Iwasaki H, Nakamura H, Shinagawa H, Morikawa K. Atomic structure of the RuvC resolvase: a Holliday junction-specific endonuclease from E. coli. Cell. 1994;78:1063–1072. - PubMed
-
- Bennett R J, Dunderdale H J, West S C. Resolution of Holliday junctions by RuvC resolvase: cleavage specificity and DNA distortion. Cell. 1993;74:1021–1031. - PubMed
-
- Bennett R J, West S C. Structural analysis of the RuvC-Holliday junction complex reveals an unfolded junction. J Mol Biol. 1995;252:213–226. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

