HP0333, a member of the dprA family, is involved in natural transformation in Helicobacter pylori
- PMID: 10482496
- PMCID: PMC94075
- DOI: 10.1128/JB.181.18.5572-5580.1999
HP0333, a member of the dprA family, is involved in natural transformation in Helicobacter pylori
Abstract
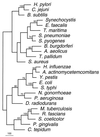
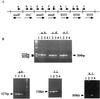
Helicobacter pylori is naturally competent for DNA transformation, but the mechanism by which transformation occurs is not known. For Haemophilus influenzae, dprA is required for transformation by chromosomal but not plasmid DNA, and the complete genomic sequence of H. pylori 26695 revealed a dprA homolog (HP0333). Examination of genetic databases indicates that DprA homologs are present in a wide variety of bacterial species. To examine whether HP0333 has a function similar to dprA of H. influenzae, HP0333, present in each of 11 strains studied, was disrupted in two H. pylori isolates. For both mutants, the frequency of transformation by H. pylori chromosomal DNA was markedly reduced, but not eliminated, compared to their wild-type parental strains. Mutation of HP0333 also resulted in a marked decrease in transformation frequency by a shuttle plasmid (pHP1), which differs from the phenotype described in H. influenzae. Complementation of the mutant with HP0333 inserted in trans in the chromosomal ureAB locus completely restored the frequency of transformation to that of the wild-type strain. Thus, while dprA is required for high-frequency transformation, transformation also may occur independently of DprA. The presence of DprA homologs in bacteria known not to be naturally competent suggests a broad function in DNA processing.
Figures



References
-
- Advanced Center for Genome Technology, University of Oklahoma. 23 July 1999, revision date. [Online.] http://www.genome.ou.edu/. [30 July 1999, last date accessed.]
-
- Alm R A, Ling L L, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Nickelsen M U, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

