Inactivation and regulation of the aerobic C(4)-dicarboxylate transport (dctA) gene of Escherichia coli
- PMID: 10482502
- PMCID: PMC94081
- DOI: 10.1128/JB.181.18.5624-5635.1999
Inactivation and regulation of the aerobic C(4)-dicarboxylate transport (dctA) gene of Escherichia coli
Abstract
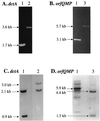
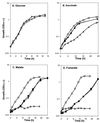
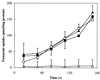
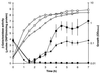
The gene (dctA) encoding the aerobic C(4)-dicarboxylate transporter (DctA) of Escherichia coli was previously mapped to the 79-min region of the linkage map. The nucleotide sequence of this region reveals two candidates for the dctA gene: f428 at 79.3 min and the o157a-o424-o328 (or orfQMP) operon at 79.9 min. The f428 gene encodes a homologue of the Sinorhizobium meliloti and Rhizobium leguminosarum H(+)/C(4)-dicarboxylate symporter, DctA, whereas the orfQMP operon encodes homologues of the aerobic periplasmic-binding protein- dependent C(4)-dicarboxylate transport system (DctQ, DctM, and DctP) of Rhodobacter capsulatus. To determine which, if either, of these loci specify the E. coli DctA system, the chromosomal f428 and orfM genes were inactivated by inserting Sp(r) or Ap(r) cassettes, respectively. The resulting f428 mutant was unable to grow aerobically with fumarate or malate as the sole carbon source and grew poorly with succinate. Furthermore, fumarate uptake was abolished in the f428 mutant and succinate transport was approximately 10-fold lower than that of the wild type. The growth and fumarate transport deficiencies of the f428 mutant were complemented by transformation with an f428-containing plasmid. No growth defect was found for the orfM mutant. In combination, the above findings confirm that f428 corresponds to the dctA gene and indicate that the orfQMP products play no role in C(4)-dicarboxylate transport. Regulation studies with a dctA-lacZ (f428-lacZ) transcriptional fusion showed that dctA is subject to cyclic AMP receptor protein (CRP)-dependent catabolite repression and ArcA-mediated anaerobic repression and is weakly induced by the DcuS-DcuR system in response to C(4)-dicarboxylates and citrate. Interestingly, in a dctA mutant, expression of dctA is constitutive with respect to C(4)-dicarboxylate induction, suggesting that DctA regulates its own synthesis. Northern blot analysis revealed a single, monocistronic dctA transcript and confirmed that dctA is subject to regulation by catabolite repression and CRP. Reverse transcriptase-mediated primer extension indicated a single transcriptional start site centered 81 bp downstream of a strongly predicted CRP-binding site.
Figures


Similar articles
-
Identification and characterization of a two-component sensor-kinase and response-regulator system (DcuS-DcuR) controlling gene expression in response to C4-dicarboxylates in Escherichia coli.J Bacteriol. 1999 Feb;181(4):1238-48. doi: 10.1128/JB.181.4.1238-1248.1999. J Bacteriol. 1999. PMID: 9973351 Free PMC article.
-
Regulation of Aerobic Succinate Transporter dctA of E. coli by cAMP-CRP, DcuS-DcuR, and EIIAGlc: Succinate as a Carbon Substrate and Signaling Molecule.Microb Physiol. 2024;34(1):108-120. doi: 10.1159/000538095. Epub 2024 Mar 1. Microb Physiol. 2024. PMID: 38432210
-
TRAP transporters: a new family of periplasmic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse gram-negative bacteria.J Bacteriol. 1997 Sep;179(17):5482-93. doi: 10.1128/jb.179.17.5482-5493.1997. J Bacteriol. 1997. PMID: 9287004 Free PMC article.
-
Cooperation of Secondary Transporters and Sensor Kinases in Transmembrane Signalling: The DctA/DcuS and DcuB/DcuS Sensor Complexes of Escherichia coli.Adv Microb Physiol. 2016;68:139-67. doi: 10.1016/bs.ampbs.2016.02.003. Epub 2016 Mar 16. Adv Microb Physiol. 2016. PMID: 27134023 Review.
-
C4-dicarboxylate carriers and sensors in bacteria.Biochim Biophys Acta. 2002 Jan 17;1553(1-2):39-56. doi: 10.1016/s0005-2728(01)00233-x. Biochim Biophys Acta. 2002. PMID: 11803016 Review.
Cited by
-
Horizontal gene transfer and the evolution of transcriptional regulation in Escherichia coli.Genome Biol. 2008 Jan 7;9(1):R4. doi: 10.1186/gb-2008-9-1-r4. Genome Biol. 2008. PMID: 18179685 Free PMC article.
-
A stress-induced block in dicarboxylate uptake and utilization in Salmonella.J Bacteriol. 2021 May 1;203(9):e00487-20. doi: 10.1128/JB.00487-20. Epub 2021 Feb 16. J Bacteriol. 2021. PMID: 33593945 Free PMC article.
-
Responsiveness of Aromatoleum aromaticum EbN1T to Lignin-Derived Phenylpropanoids.Appl Environ Microbiol. 2021 May 11;87(11):e03140-20. doi: 10.1128/AEM.03140-20. Print 2021 May 11. Appl Environ Microbiol. 2021. PMID: 33741621 Free PMC article.
-
Positive Selection during Niche Adaptation Results in Large-Scale and Irreversible Rearrangement of Chromosomal Gene Order in Bacteria.Mol Biol Evol. 2022 Apr 10;39(4):msac069. doi: 10.1093/molbev/msac069. Mol Biol Evol. 2022. PMID: 35348727 Free PMC article.
-
Tetracarboxylic acid transporter regulates growth, conidiation, and carbon utilization in Metarhizium acridum.Appl Microbiol Biotechnol. 2023 May;107(9):2969-2982. doi: 10.1007/s00253-023-12471-x. Epub 2023 Mar 21. Appl Microbiol Biotechnol. 2023. PMID: 36941435
References
-
- Aiba H, Adkya S, de Crombrugghe B. Evidence for a functional gal promoter in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–11910. - PubMed
-
- Bewick M A, Lo T C Y. Dicarboxylic acid transport in Escherichia coli K12: involvement of a binding protein in the translocation of dicarboxylic acids across the outer membrane of the cell envelope. Can J Biochem. 1979;57:653–661. - PubMed
-
- Bewick M A, Lo T C Y. Localization of the dicarboxylate binding protein in the cell envelope of Escherichia coli K12. Can J Biochem. 1980;58:885–897. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

