Genomic subtraction identifies Salmonella typhimurium prophages, F-related plasmid sequences, and a novel fimbrial operon, stf, which are absent in Salmonella typhi
- PMID: 10482505
- PMCID: PMC94084
- DOI: 10.1128/JB.181.18.5652-5661.1999
Genomic subtraction identifies Salmonella typhimurium prophages, F-related plasmid sequences, and a novel fimbrial operon, stf, which are absent in Salmonella typhi
Abstract
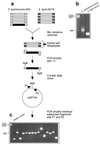
Salmonella typhimurium causes systemic and fatal infection in inbred mice, while the related serotype Salmonella typhi is avirulent for mammals other than humans. In order to identify genes from the virulent strain S. typhimurium ATCC 14028 that are absent in S. typhi Ty2, and therefore might be involved in S. typhimurium mouse virulence, a PCR-supported genomic subtractive hybridization procedure was employed. We have identified a novel putative fimbrial operon, stfACDEFG, located at centisome 5 of the S. typhimurium chromosome, which is absent in S. typhi, Salmonella arizonae, and Salmonella bongori but was detected in several other Salmonella serotypes. The fimbrial genes represent a genomic insertion in S. typhimurium compared to the respective region between fhuB and hemL in Escherichia coli K-12. In addition, the subtraction procedure yielded F plasmid-related sequences from the S. typhimurium virulence plasmid, a number of DNA fragments representing parts of lambdoid prophages and putative sugar transporters, and several fragments with unknown sequences. The majority of subtracted chromosomal sequences map to three distinct locations, around centisomes 5, 27, and 57.
Figures








References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

