Replacement of the bacteriophage Mu strong gyrase site and effect on Mu DNA replication
- PMID: 10482521
- PMCID: PMC94100
- DOI: 10.1128/JB.181.18.5783-5789.1999
Replacement of the bacteriophage Mu strong gyrase site and effect on Mu DNA replication
Abstract
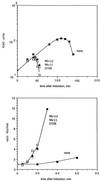
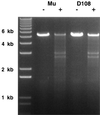
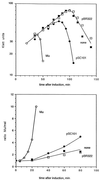
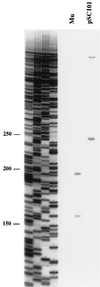
The bacteriophage Mu strong gyrase site (SGS) is required for efficient replicative transposition and functions by promoting the synapsis of prophage termini. To look for other sites which could substitute for the SGS in promoting Mu replication, we have replaced the SGS in the middle of the Mu genome with fragments of DNA from various sources. A central fragment from the transposing virus D108 allowed efficient Mu replication and was shown to contain a strong gyrase site. However, neither the strong gyrase site from the plasmid pSC101 nor the major gyrase site from pBR322 could promote efficient Mu replication, even though the pSC101 site is a stronger gyrase site than the Mu SGS as assayed by cleavage in the presence of gyrase and the quinolone enoxacin. To look for SGS-like sites in the Escherichia coli chromosome which might be involved in organizing nucleoid structure, fragments of E. coli chromosomal DNA were substituted for the SGS: first, repeat sequences associated with gyrase binding (bacterial interspersed mosaic elements), and, second, random fragments of the entire chromosome. No fragments were found that could replace the SGS in promoting efficient Mu replication. These results demonstrate that the gyrase sites from the transposing phages possess unusual properties and emphasize the need to determine the basis of these properties.
Figures


Similar articles
-
Genetic analysis of the strong gyrase site (SGS) of bacteriophage Mu: localization of determinants required for promoting Mu replication.Mol Microbiol. 2000 Aug;37(4):800-10. doi: 10.1046/j.1365-2958.2000.02042.x. Mol Microbiol. 2000. PMID: 10972802
-
A biochemical analysis of the interaction of DNA gyrase with the bacteriophage Mu, pSC101 and pBR322 strong gyrase sites: the role of DNA sequence in modulating gyrase supercoiling and biological activity.Mol Microbiol. 2003 Oct;50(1):333-47. doi: 10.1046/j.1365-2958.2003.03690.x. Mol Microbiol. 2003. PMID: 14507384
-
The Mu strong gyrase-binding site promotes efficient synapsis of the prophage termini.Mol Microbiol. 1996 Oct;22(2):283-92. doi: 10.1046/j.1365-2958.1996.00115.x. Mol Microbiol. 1996. PMID: 8930913
-
Transposable Phage Mu.Microbiol Spectr. 2014 Oct;2(5):10.1128/microbiolspec.MDNA3-0007-2014. doi: 10.1128/microbiolspec.MDNA3-0007-2014. Microbiol Spectr. 2014. PMID: 26104374 Free PMC article. Review.
-
Mechanism of bacteriophage mu transposition.Annu Rev Genet. 1986;20:385-429. doi: 10.1146/annurev.ge.20.120186.002125. Annu Rev Genet. 1986. PMID: 3028246 Review. No abstract available.
Cited by
-
Measuring chromosome dynamics on different time scales using resolvases with varying half-lives.Mol Microbiol. 2005 May;56(4):1049-61. doi: 10.1111/j.1365-2958.2005.04588.x. Mol Microbiol. 2005. PMID: 15853889 Free PMC article.
-
A cell engineering strategy to enhance supercoiled plasmid DNA production for gene therapy.Biotechnol Bioeng. 2016 Sep;113(9):2064-71. doi: 10.1002/bit.25971. Epub 2016 Mar 16. Biotechnol Bioeng. 2016. PMID: 26928284 Free PMC article.
-
Single-nucleotide-resolution mapping of DNA gyrase cleavage sites across the Escherichia coli genome.Nucleic Acids Res. 2019 Feb 20;47(3):1373-1388. doi: 10.1093/nar/gky1222. Nucleic Acids Res. 2019. PMID: 30517674 Free PMC article.
-
Application of Plasmid Engineering to Enhance Yield and Quality of Plasmid for Vaccine and Gene Therapy.Bioengineering (Basel). 2019 Jun 19;6(2):54. doi: 10.3390/bioengineering6020054. Bioengineering (Basel). 2019. PMID: 31248216 Free PMC article.
-
Mu-like prophage strong gyrase site sequences: analysis of properties required for promoting efficient mu DNA replication.J Bacteriol. 2004 Jul;186(14):4575-84. doi: 10.1128/JB.186.14.4575-4584.2004. J Bacteriol. 2004. PMID: 15231790 Free PMC article.
References
-
- Aldaz H, Schuster E, Baker T A. The interwoven architecture of the Mu transposase couples DNA synapsis to catalysis. Cell. 1996;85:257–269. - PubMed
-
- Bachellier S, Gilsen E, Hofnung M, Hill C W. Repeated sequences. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznickoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: American Society for Microbiology; 1996. pp. 2012–2040.
-
- Baxa C A, Chiang L, Howe M M. DNA sequence characterization of the G gene region of bacteriophage Mu. J DNA Sequencing Mapping. 1992;2:329–333. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

