Mutagenesis of the NS2B-NS3-mediated cleavage site in the flavivirus capsid protein demonstrates a requirement for coordinated processing
- PMID: 10482557
- PMCID: PMC112824
- DOI: 10.1128/JVI.73.10.8083-8094.1999
Mutagenesis of the NS2B-NS3-mediated cleavage site in the flavivirus capsid protein demonstrates a requirement for coordinated processing
Abstract
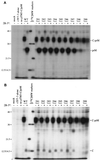
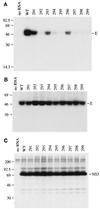
Analysis of flavivirus polyprotein processing has revealed the presence of a substrate for the virus-encoded NS2B-NS3 protease at the carboxy-terminal end of the C (capsid or core) protein. Cleavage at this site has been implicated in the efficient generation of the amino terminus of prM via signal peptidase cleavage. Yellow fever virus has four basic residues (Arg-Lys-Arg-Arg) in the P1 through P4 positions of this cleavage site. Multiple alanine substitutions were made for these residues in order to investigate the substrate specificity and biological significance of this cleavage. Mutants were analyzed by several methods: (i) a cell-free trans processing assay for direct analysis of NS2B-NS3-mediated cleavage; (ii) a trans processing assay in BHK-21 cells, using a C-prM polyprotein, for analysis of prM production; (iii) an infectivity assay of full-length transcripts to determine plaque-forming ability; and (iv) analysis of proteins expressed from full-length transcripts to assess processing in the context of the complete genome. Mutants that exhibited severe defects in processing in vitro and in vivo were incapable of forming plaques. Mutants that contained two adjacent basic residues within the P1 through P4 region were processed more efficiently in vitro and in vivo, and transcripts bearing these mutations were fully infectious. Furthermore, two naturally occurring plaque-forming revertants were analyzed and shown to have restored protein processing phenotypes in vivo. Finally, the efficient production of prM was shown to be dependent on the proteolytic activity of NS3. These data support a model of two coordinated cleavages, one that generates the carboxy terminus of C and another that generates the amino terminus of prM. A block in the viral protease-mediated cleavage inhibits the production of prM by the signal peptidase, inhibits particle release, and eliminates plaque formation.
Figures



References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1998.
-
- Barrett A J. Classification of peptidases. Methods Enzymol. 1994;244:1–15. - PubMed
-
- Bredenbeek, P. J., E. Kooi, B. D. Lindenbach, M. Lucassen, N. Huijkman, W. J. M. Spaan, and C. M. Rice. Unpublished data.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

