The first immunoglobulin-like domain of HveC is sufficient to bind herpes simplex virus gD with full affinity, while the third domain is involved in oligomerization of HveC
- PMID: 10482562
- PMCID: PMC112829
- DOI: 10.1128/JVI.73.10.8127-8137.1999
The first immunoglobulin-like domain of HveC is sufficient to bind herpes simplex virus gD with full affinity, while the third domain is involved in oligomerization of HveC
Abstract
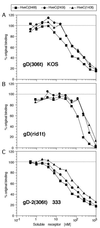
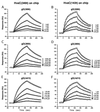
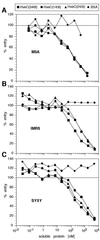
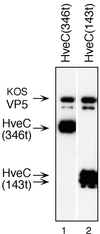
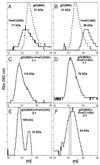
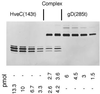
The human herpesvirus entry mediator C (HveC/PRR1) is a member of the immunoglobulin family used as a cellular receptor by the alphaherpesviruses herpes simplex virus (HSV), pseudorabies virus, and bovine herpesvirus type 1. We previously demonstrated direct binding of the purified HveC ectodomain to purified HSV type 1 (HSV-1) and HSV-2 glycoprotein D (gD). Here, using a baculovirus expression system, we constructed and purified truncated forms of the receptor containing one [HveC(143t)], two [HveC(245t)], or all three immunoglobulin-like domains [HveC(346t)] of the extracellular region. All three constructs were equally able to compete with HveC(346t) for gD binding. The variable domain bound to virions and blocked HSV infection as well as HveC(346t). Thus, all of the binding to the receptor occurs within the first immunoglobulin-like domain, or V-domain, of HveC. These data confirm and extend those of Cocchi et al. (F. Cocchi, M. Lopez, L. Menotti, M. Aoubala, P. Dubreuil, and G. Campadelli-Fiume, Proc. Natl. Acad. Sci. USA 95:15700, 1998). Using biosensor analysis, we measured the affinity of binding of gD from HSV strains KOS and rid1 to two forms of HveC. Soluble gDs from the KOS strain of HSV-1 had the same affinity for HveC(346t) and HveC(143t). The mutant gD(rid1t) had an increased affinity for HveC(346t) and HveC(143t) due to a faster rate of complex formation. Interestingly, we found that HveC(346t) was a tetramer in solution, whereas HveC(143t) and HveC(245t) formed dimers, suggesting a role for the third immunoglobulin-like domain of HveC in oligomerization. In addition, the stoichiometry between gD and HveC appeared to be influenced by the level of HveC oligomerization.
Figures



Similar articles
-
Glycoprotein D homologs in herpes simplex virus type 1, pseudorabies virus, and bovine herpes virus type 1 bind directly to human HveC(nectin-1) with different affinities.Virology. 2001 Feb 1;280(1):7-18. doi: 10.1006/viro.2000.0747. Virology. 2001. PMID: 11162814
-
Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry.J Virol. 1998 Sep;72(9):7064-74. doi: 10.1128/JVI.72.9.7064-7074.1998. J Virol. 1998. PMID: 9696799 Free PMC article.
-
Use of chimeric nectin-1(HveC)-related receptors to demonstrate that ability to bind alphaherpesvirus gD is not necessarily sufficient for viral entry.Virology. 2001 Jul 5;285(2):366-75. doi: 10.1006/viro.2001.0989. Virology. 2001. PMID: 11437670
-
The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells.Rev Med Virol. 2000 Sep-Oct;10(5):305-19. doi: 10.1002/1099-1654(200009/10)10:5<305::aid-rmv286>3.0.co;2-t. Rev Med Virol. 2000. PMID: 11015742 Review.
-
The role of herpes simplex virus glycoproteins in the virus replication cycle.Acta Virol. 1998 Apr;42(2):103-18. Acta Virol. 1998. PMID: 9770079 Review.
Cited by
-
The herpes simplex virus JMP mutant enters receptor-negative J cells through a novel pathway independent of the known receptors nectin1, HveA, and nectin2.J Virol. 2004 May;78(9):4720-9. doi: 10.1128/jvi.78.9.4720-4729.2004. J Virol. 2004. PMID: 15078954 Free PMC article.
-
Immunoglobulin superfamily virus receptors and the evolution of adaptive immunity.PLoS Pathog. 2009 Nov;5(11):e1000481. doi: 10.1371/journal.ppat.1000481. Epub 2009 Nov 26. PLoS Pathog. 2009. PMID: 19956667 Free PMC article. Review. No abstract available.
-
Displacement of the C terminus of herpes simplex virus gD is sufficient to expose the fusion-activating interfaces on gD.J Virol. 2013 Dec;87(23):12656-66. doi: 10.1128/JVI.01727-13. Epub 2013 Sep 18. J Virol. 2013. PMID: 24049165 Free PMC article.
-
Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor.Sci Adv. 2020 May 13;6(20):eaba5147. doi: 10.1126/sciadv.aba5147. eCollection 2020 May. Sci Adv. 2020. PMID: 32426511 Free PMC article.
-
Chronic lymphocytic leukaemia with necrotic herpetic adenitis: an elusive clinical condition.BMJ Case Rep. 2018 Feb 2;2018:bcr2017222091. doi: 10.1136/bcr-2017-222091. BMJ Case Rep. 2018. PMID: 29420244 Free PMC article.
References
-
- Aoki J, Koike S, Asou H, Ise I, Suwa H, Tanaka T, Miyasaka M, Nomoto A. Mouse homolog of poliovirus receptor-related gene 2 product, mPRR2, mediates homophilic cell aggregation. Exp Cell Res. 1997;235:374–384. - PubMed
-
- Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. - PubMed
-
- Biacore Inc. BIAevaluation software handbook, version 3.0. Uppsala, Sweden: Biacore AB; 1997.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

