Evidence of recombination among enteroviruses
- PMID: 10482628
- PMCID: PMC112895
- DOI: 10.1128/JVI.73.10.8741-8749.1999
Evidence of recombination among enteroviruses
Abstract
Human enteroviruses consist of more than 60 serotypes, reflecting a wide range of evolutionary divergence. They have been genetically classified into four clusters on the basis of sequence homology in the coding region of the single-stranded RNA genome. To explore further the genetic relationships between human enteroviruses and to characterize the evolutionary mechanisms responsible for variation, previously sequenced genomes were subjected to detailed comparison. Bootstrap and genetic similarity analyses were used to systematically scan the alignments of complete genomic sequences. Bootstrap analysis provided evidence from an early recombination event at the junction of the 5' noncoding and coding regions of the progenitors of the current clusters. Analysis within the genetic clusters indicated that enterovirus prototype strains include intraspecies recombinants. Recombination breakpoints were detected in all genomic regions except the capsid protein coding region. Our results suggest that recombination is a significant and relatively frequent mechanism in the evolution of enterovirus genomes.
Figures
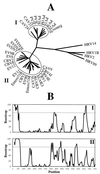
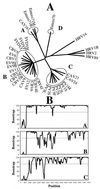
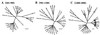

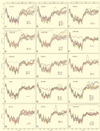

References
-
- Cammack N, Phillips A, Dunn G, Patel V, Minor P D. Intertypic genomic rearrangements of poliovirus strains in vaccinees. Virology. 1988;167:507–514. - PubMed
-
- Carr J K, Salminen M O, Albert J, Sanders-Buell E, Gotte D, Birx D L, McCutchan F E. Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology. 1998;247:22–31. - PubMed
-
- Evans D J, Almond J W. Cell receptors for picornaviruses as determinants of cell tropism and pathogenesis. Trends Microbiol. 1998;6:198–202. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

