Components of an SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication cycle
- PMID: 10485847
- PMCID: PMC316987
- DOI: 10.1101/gad.13.17.2242
Components of an SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication cycle
Abstract
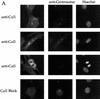
Centrosomes organize the mitotic spindle to ensure accurate segregation of the chromosomes in mitosis. The mechanism that ensures accurate duplication and separation of the centrosomes underlies the fidelity of chromosome segregation, but remains unknown. In Saccharomyces cerevisiae, entry into S phase and separation of spindle pole bodies each require CDC4 and CDC34, which encode components of an SCF (Skp1-cullin-F-box) ubiquitin ligase, but a direct (SCF) connection to the spindle pole body is unknown. Using immunofluorescence microscopy, we show that in mammalian cells the Skp1 protein and the cullin Cul1 are localized to interphase and mitotic centrosomes and to the cytoplasm and nucleus. Deconvolution and immunoelectron microscopy suggest that Skp1 forms an extended pericentriolar structure that may function to organize the centrosome. Purified centrosomes also contain Skp1, and Cul1 modified by the ubiquitin-like molecule NEDD8, suggesting a role for NEDD8 in targeting. Using an in vitro assay for centriole separation in Xenopus extracts, antibodies to Skp1 or Cul1 block separation. Proteasome inhibitors block both centriole separation in vitro and centrosome duplication in Xenopus embryos. We identify candidate centrosomal F-box proteins, suggesting that distinct SCF complexes may direct proteolysis of factors mediating multiple steps in the centrosome cycle.
Figures







Similar articles
-
Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34.Genes Dev. 1999 Jun 15;13(12):1614-26. doi: 10.1101/gad.13.12.1614. Genes Dev. 1999. PMID: 10385629 Free PMC article.
-
The SCF(HOS/beta-TRCP)-ROC1 E3 ubiquitin ligase utilizes two distinct domains within CUL1 for substrate targeting and ubiquitin ligation.Mol Cell Biol. 2000 Feb;20(4):1382-93. doi: 10.1128/MCB.20.4.1382-1393.2000. Mol Cell Biol. 2000. PMID: 10648623 Free PMC article.
-
Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1.Genes Dev. 1998 Aug 15;12(16):2587-97. doi: 10.1101/gad.12.16.2587. Genes Dev. 1998. PMID: 9716410 Free PMC article.
-
Proteolysis and the G1-S transition: the SCF connection.Curr Opin Genet Dev. 1998 Feb;8(1):36-42. doi: 10.1016/s0959-437x(98)80059-2. Curr Opin Genet Dev. 1998. PMID: 9529603 Review.
-
Role of SKP1-CUL1-F-box-protein (SCF) E3 ubiquitin ligases in skin cancer.J Genet Genomics. 2013 Mar 20;40(3):97-106. doi: 10.1016/j.jgg.2013.02.001. Epub 2013 Feb 10. J Genet Genomics. 2013. PMID: 23522382 Free PMC article. Review.
Cited by
-
Dynamic relocalization of histone MacroH2A1 from centrosomes to inactive X chromosomes during X inactivation.J Cell Biol. 2000 Sep 4;150(5):1189-98. doi: 10.1083/jcb.150.5.1189. J Cell Biol. 2000. PMID: 10974005 Free PMC article.
-
Mutational analysis reveals a role for the C terminus of the proteasome subunit Rpt4p in spindle pole body duplication in Saccharomyces cerevisiae.Genetics. 2002 Oct;162(2):705-20. doi: 10.1093/genetics/162.2.705. Genetics. 2002. PMID: 12399382 Free PMC article.
-
Fbxo41 Promotes Disassembly of Neuronal Primary Cilia.Sci Rep. 2019 Jun 3;9(1):8179. doi: 10.1038/s41598-019-44589-2. Sci Rep. 2019. PMID: 31160656 Free PMC article.
-
SCF ubiquitin ligases in the maintenance of genome stability.Trends Biochem Sci. 2012 Feb;37(2):66-73. doi: 10.1016/j.tibs.2011.10.004. Epub 2011 Nov 16. Trends Biochem Sci. 2012. PMID: 22099186 Free PMC article.
-
SCF E3-mediated autoubiquitination negatively regulates activity of Cdc34 E2 but plays a nonessential role in the catalytic cycle in vitro and in vivo.Mol Cell Biol. 2007 Aug;27(16):5860-70. doi: 10.1128/MCB.01555-06. Epub 2007 Jun 11. Mol Cell Biol. 2007. PMID: 17562869 Free PMC article.
References
-
- Agard DA, Hiraoka Y, Shaw P, Sedat JW. Fluorescence microscopy in three dimensions. In: Taylor DL, Wang Y-l, editors. Fluorescence microscopy of living cells in culture. San Diego, CA: Academic Press; 1989. pp. 353–377. - PubMed
-
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. - PubMed
-
- Bornens M, Paintrand M, Berges J, Marty M-C, Karsenti E. Structural and chemical characterization of isolated centrosomes. Cell Mobility Cytoskeleton. 1987;8:238–249. - PubMed
-
- Bouckson-Castaing V, Moudjou M, Ferguson DJ, Mucklow S, Belkaid Y, Milon G, Crocker PR. Molecular characterization of ninein, a new coiled-coil protein of the centrosome. J Cell Sci. 1996;109:179–190. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
