Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4
- PMID: 10485849
- PMCID: PMC316991
- DOI: 10.1101/gad.13.17.2271
Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4
Abstract
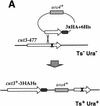
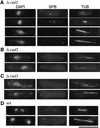
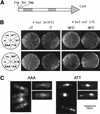
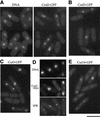
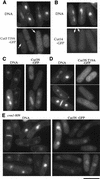
The condensin complex in frog extracts, containing two SMC (structural maintenance of chromosomes) and three non-SMC subunits, promotes mitotic chromosome condensation, and its supercoiling activity increases during mitosis by Cdc2 phosphorylation. Here, we report that fission yeast has the same five-member condensin complex, each of which is essential for mitotic condensation. The condensin complex was purified and the subunits were identified by microsequencing. Cnd1, Cnd2, and Cnd3, three non-SMC subunits showing a high degree of sequence conservation to frog subunits, are essential for viability, and their gene disruption leads to a phenotype indistinguishable from that observed in cut3-477 and cut14-208, known mutations in SMC4 and SMC2-like subunits. Condensin subunits tagged with GFP were observed to alter dramatically their localization during the cell cycle, enriched in the nucleus during mitosis, but cytoplasmic during other stages. This stage-specific alteration in localization requires mitosis-specific phosphorylation of the T19 Cdc2 site in Cut3. The T19 site is phosphorylated in vitro by Cdc2 kinase and shows the maximal phosphorylation in metaphase in vivo. Its alanine substitution mutant fails to suppress the temperature-sensitive phenotype of cut3-477, and shows deficiency in condensation, probably because Cut3 T19A remains cytoplasmic. Therefore, direct Cdc2 phosphorylation of fission yeast condensin may facilitate its nuclear accumulation during mitosis.
Figures









References
-
- Adachi Y, Luke M, Laemmli UK. Chromosome assembly in vitro: Topoisomerase II is required for condensation. Cell. 1991;64:137–148. - PubMed
-
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. - PubMed
-
- Bhat MA, Philp AV, Glover DM, Bellen HJ. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with Topoisomerase II. Cell. 1996;87:1103–1114. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
