Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation
- PMID: 10485883
- PMCID: PMC17940
- DOI: 10.1073/pnas.96.19.10667
Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation
Abstract
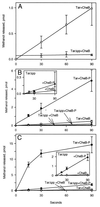
The mechanistic basis of sensory adaptation and gradient sensing in bacterial chemotaxis is reversible covalent modification of transmembrane chemoreceptors, methylation, and demethylation at specific glutamyl residues in their cytoplasmic domains. These reactions are catalyzed by a dedicated methyltransferase CheR and a dedicated methylesterase CheB. The esterase is also a deamidase that creates certain methyl-accepting glutamyls by hydrolysis of glutamine side chains. We investigated the action of CheB and its activated form, phospho-CheB, on a truncated form of the aspartate receptor of Escherichia coli that was missing the last 5 aa of the intact receptor. The deleted pentapeptide is conserved in several chemoreceptors in enteric and related bacteria. The truncated receptor was much less efficiently demethylated and deamidated than intact receptor, but essentially was unperturbed for kinase activation or transmembrane signaling. CheB bound specifically to an affinity column carrying the isolated pentapeptide, implying that in the intact receptor the pentapeptide serves as a docking site for the methylesterase/deamidase and that the truncated receptor was inefficiently modified because the enzyme could not dock. It is striking that the same pentapeptide serves as an activity-enhancing docking site for the methyltransferase CheR, the other enzyme involved in adaptational covalent modification of chemoreceptors. A shared docking site raises the tantalizing possibility that relative rates of methylation and demethylation could be influenced by competition between the two enzymes at that site.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

