Modeling evolutionary landscapes: mutational stability, topology, and superfunnels in sequence space
- PMID: 10485887
- PMCID: PMC17944
- DOI: 10.1073/pnas.96.19.10689
Modeling evolutionary landscapes: mutational stability, topology, and superfunnels in sequence space
Abstract
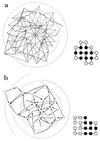
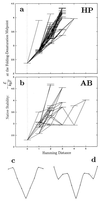
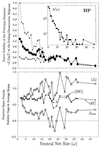
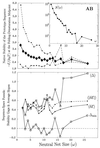
Random mutations under neutral or near-neutral conditions are studied by considering plausible evolutionary trajectories on "neutral nets"-i.e., collections of sequences (genotypes) interconnected via single-point mutations encoding for the same ground-state structure (phenotype). We use simple exact lattice models for the mapping between sequence and conformational spaces. Densities of states based on model intrachain interactions are determined by exhaustive conformational enumeration. We compare results from two very different interaction schemes to ascertain robustness of the conclusions. In both models, sequences in a majority of neutral nets center around a single "prototype sequence" of maximum mutational stability, tolerating the largest number of neutral mutations. General analytical considerations show that these topologies by themselves lead to higher steady-state evolutionary populations at prototype sequences. On average, native thermodynamic stability increases toward a maximum at the prototype sequence, resulting in funnel-like arrangements of native stabilities in sequence space. These observations offer a unified perspective on sequence design, native stability, and mutational stability of proteins. These principles are generalizable from native stability to any measure of fitness provided that its variation with respect to mutations is essentially smooth.
Figures
References
-
- Wright S. In: Proceedings of the Sixth International Congress on Genetics, Vol. 1. Jones D F, editor. New York: Brooklyn Botanic Gardens; 1932. pp. 356–366.
-
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge: Cambridge Univ. Press; 1983.
-
- Kauffman S, Levin S. J Theor Biol. 1987;128:11–45. - PubMed
-
- Fontana W, Schuster P. Biophys Chem. 1987;26:123–147. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

