Capillary electrophoresis-single-strand conformation polymorphism analysis for rapid identification of Pseudomonas aeruginosa and other gram-negative nonfermenting bacilli recovered from patients with cystic fibrosis
- PMID: 10488211
- PMCID: PMC85573
- DOI: 10.1128/JCM.37.10.3374-3379.1999
Capillary electrophoresis-single-strand conformation polymorphism analysis for rapid identification of Pseudomonas aeruginosa and other gram-negative nonfermenting bacilli recovered from patients with cystic fibrosis
Abstract
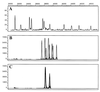
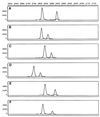
We used capillary electrophoresis-single-strand conformation polymorphism (CE-SSCP) analysis of PCR-amplified 16S rRNA gene fragments for rapid identification of Pseudomonas aeruginosa and other gram-negative nonfermenting bacilli isolated from patients with cystic fibrosis (CF). Target sequences were amplified by using forward and reverse primers labeled with various fluorescent dyes. The labeled PCR products were denatured by heating and separated by capillary gel electrophoresis with an automated DNA sequencer. Data were analyzed with GeneScan 672 software. This program made it possible to control lane-to-lane variability by standardizing the peak positions relative to internal DNA size markers. Thirty-four reference strains belonging to the genera Pseudomonas, Brevundimonas, Burkholderia, Comamonas, Ralstonia, Stenotrophomonas, and Alcaligenes were tested with primer sets spanning 16S rRNA gene regions with various degrees of polymorphism. The best results were obtained with the primer set P11P-P13P, which spans a moderately polymorphic region (Escherichia coli 16S rRNA positions 1173 to 1389 [M. N. Widjojoatmodjo, A. C. Fluit, and J. Verhoef, J. Clin. Microbiol. 32:3002-3007, 1994]). This primer set differentiated the main CF pathogens from closely related species but did not distinguish P. aeruginosa from Pseudomonas alcaligenes-Pseudomonas pseudoalcaligenes and Alcaligenes xylosoxidans from Alcaligenes denitrificans. Two hundred seven CF clinical isolates (153 of P. aeruginosa, 26 of Stenotrophomonas maltophilia, 15 of Burkholderia spp., and 13 of A. xylosoxidans) were tested with P11P-P13P. The CE-SSCP patterns obtained were identical to those for the corresponding reference strains. Fluorescence-based CE-SSCP analysis is simple to use, gives highly reproducible results, and makes it possible to analyze a large number of strains. This approach is suited for the rapid identification of the main gram-negative nonfermenting bacilli encountered in CF.
Figures
References
-
- Arakawa H, Tsuji A, Maeda M, Kamahori M, Kambara H. Analysis of single-strand conformation polymorphisms by capillary electrophoresis with laser induced fluorescence detection. J Pharm Biomed Anal. 1997;15:1537–1544. - PubMed
-
- Burdge D R, Noble M A, Campbell M E, Krell V L, Speert D P. Xanthomonas maltophilia misidentified as Pseudomonas cepacia in cultures of sputum from patients with cystic fibrosis: a diagnostic pitfall with major clinical implications. Clin Infect Dis. 1995;20:445–448. - PubMed
-
- Dunne W M J, Maisch S. Epidemiological investigation of infections due to Alcaligenes species in children and patients with cystic fibrosis: use of repetitive-element-sequence polymerase chain reaction. Clin Infect Dis. 1995;20:836–841. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

