Identification of a novel dexamethasone-sensitive RNA-destabilizing region on rat monocyte chemoattractant protein 1 mRNA
- PMID: 10490587
- PMCID: PMC84617
- DOI: 10.1128/MCB.19.10.6471
Identification of a novel dexamethasone-sensitive RNA-destabilizing region on rat monocyte chemoattractant protein 1 mRNA
Abstract
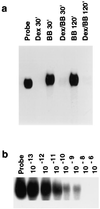
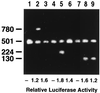
Glucocorticoids are potent anti-inflammatory agents widely used in the treatment of human disease. We have previously shown that the inflammatory cytokine monocyte chemoattractant protein 1 (MCP-1) is regulated posttranscriptionally by glucocorticoids in arterial smooth muscle cells (SMC). To elucidate the mechanism mediating this effect, in vitro-transcribed radiolabeled MCP-1 mRNA was incubated with cytoplasmic extracts from SMC and analyzed by gel electrophoresis. Extracts from SMC treated with platelet-derived growth factor (PDGF) did not degrade the transcripts for up to 3 h. In contrast, extracts from cells treated with 1 microM dexamethasone (Dex) alone or in combination with PDGF degraded the probe with a half-life of approximately 15 min. Dex had maximal effect at concentrations above 0.01 microM and was effective on both rat and human MCP-1 transcripts. By deletion analysis, the Dex-sensitive region of the MCP-1 mRNA was localized to the initial 224 nucleotides (nt) at the 5' end and did not involve an AU-rich sequence in the 3' untranslated end. The 224-nt region conferred Dex sensitivity to heterologous mRNA. These studies provide new insights into the molecular mechanisms underlying the effect of glucocorticoids on gene expression.
Figures






References
-
- Almawi W Y, Beyhum H N, Rahme A A, Rieder M J. Regulation of cytokine and cytokine receptor expression by glucocorticoids. J Leukoc Biol. 1996;60:563–572. - PubMed
-
- Amano Y, Lee S W, Allison A C. Inhibition by glucocorticoids of the formation of interleukin-1 alpha, interleukin-1 beta, and interleukin-6: mediation by decreased mRNA stability. Mol Pharmacol. 1993;43:176–182. - PubMed
-
- Barnes P J. Anti-inflammatory mechanisms of glucocorticoids. Biochem Soc Trans. 1995;23:940–945. - PubMed
-
- Beato M. Gene regulation by steroid hormones. Cell. 1989;56:335–344. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
