Mouse receptor interacting protein 3 does not contain a caspase-recruiting or a death domain but induces apoptosis and activates NF-kappaB
- PMID: 10490590
- PMCID: PMC84620
- DOI: 10.1128/MCB.19.10.6500
Mouse receptor interacting protein 3 does not contain a caspase-recruiting or a death domain but induces apoptosis and activates NF-kappaB
Abstract
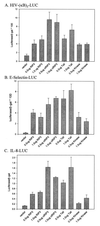
The death domain-containing receptor superfamily and their respective downstream mediators control whether or not cells initiate apoptosis or activate NF-kappaB, events critical for proper immune system function. A screen for upstream activators of NF-kappaB identified a novel serine-threonine kinase capable of activating NF-kappaB and inducing apoptosis. Based upon domain organization and sequence similarity, this novel kinase, named mRIP3 (mouse receptor interacting protein 3), appears to be a new RIP family member. RIP, RIP2, and mRIP3 contain an N-terminal kinase domain that share 30 to 40% homology. In contrast to the C-terminal death domain found in RIP or the C-terminal caspase-recruiting domain found in RIP2, the C-terminal tail of mRIP3 contains neither motif and is unique. Despite this feature, overexpression of the mRIP3 C terminus is sufficient to induce apoptosis, suggesting that mRIP3 uses a novel mechanism to induce death. mRIP3 also induced NF-kappaB activity which was inhibited by overexpression of either dominant-negative NIK or dominant-negative TRAF2. In vitro kinase assays demonstrate that mRIP3 is catalytically active and has autophosphorylation site(s) in the C-terminal domain, but the mRIP3 catalytic activity is not required for mRIP3 induced apoptosis and NF-kappaB activation. Unlike RIP and RIP2, mRIP3 mRNA is expressed in a subset of adult tissues and is thus likely to be a tissue-specific regulator of apoptosis and NF-kappaB activity. While the lack of a dominant-negative mutant precludes linking mRIP3 to a known upstream regulator, characterizing the expression pattern and the in vitro functions of mRIP3 provides insight into the mechanism(s) by which cells modulate the balance between survival and death in a cell-type-specific manner.
Figures





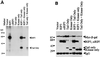

References
-
- Ashkenazi A, Dixit V M. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. - PubMed
-
- Belvin M P, Anderson K V. A conserved signaling pathway: the Drosophila toll-dorsal pathway. Ann Rev Cell Dev Biol. 1996;12:393–416. - PubMed
-
- Bodmer J L, Burns K, Schneider P, Hofmann K, Steiner V, Thome M, Bornand T, Hahne M, Schroter M, Becker K, Wilson A, French L E, Browning J L, MacDonald H R, Tschopp J. TRAMP, a novel apoptosis-mediating receptor with sequence homology to tumor necrosis factor receptor 1 and Fas(Apo-1/CD95) Immunity. 1997;6:79–88. - PubMed
-
- Chaudhary P M, Eby M, Jasmin A, Bookwalter A, Murray J, Hood L. Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NF-κB pathway. Immunity. 1997;7:821–830. - PubMed
-
- Chinnaiyan A M, O’Rourke K, Tewari M, Dixit V M. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–512. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
