Roles of cell division and gene transcription in the methylation of CpG islands
- PMID: 10490608
- PMCID: PMC84656
- DOI: 10.1128/MCB.19.10.6690
Roles of cell division and gene transcription in the methylation of CpG islands
Abstract
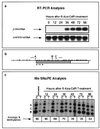
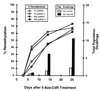
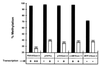
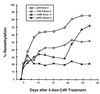
De novo methylation of CpG islands within the promoters of eukaryotic genes is often associated with their transcriptional repression, yet the methylation of CpG islands located downstream of promoters does not block transcription. We investigated the kinetics of mRNA induction, demethylation, and remethylation of the p16 promoter and second-exon CpG islands in T24 cells after 5-aza-2'-deoxycytidine (5-Aza-CdR) treatment to explore the relationship between CpG island methylation and gene transcription. The rates of remethylation of both CpG islands were associated with time but not with the rate of cell division, and remethylation of the p16 exon 2 CpG island occurred at a higher rate than that of the p16 promoter. We also examined the relationship between the remethylation of coding sequence CpG islands and gene transcription. The kinetics of remethylation of the p16 exon 2, PAX-6 exon 5, c-ABL exon 11, and MYF-3 exon 3 loci were examined following 5-Aza-CdR treatment because these genes contain exonic CpG islands which are hypermethylated in T24 cells. Remethylation occurred most rapidly in the p16, PAX-6, and c-ABL genes, shown to be transcribed prior to drug treatment. These regions also exhibited higher levels of remethylation in single-cell clones and subclones derived from 5-Aza-CdR-treated T24 cells. Our data suggest that de novo methylation is not restricted to the S phase of the cell cycle and that transcription through CpG islands does not inhibit their remethylation.
Figures


References
-
- Antequera F, Boyes J, Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;62:503–514. - PubMed
-
- Baylin S B, Herman J G, Graff J R, Vertino P M, Issa J-P. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–146. - PubMed
-
- Bender C M, Pao M M, Jones P A. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 1998;58:95–101. - PubMed
-
- Bender C M, Zingg J M, Jones P A. DNA methylation as a target for drug design. Pharmacol Res. 1998;15:175–187. - PubMed
-
- Bird A P. CpG islands as gene markers in the vertebrate nucleus. Trends Genet. 1987;3:342–347.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
