Regulation of cell cycle transcription factor Swi4 through auto-inhibition of DNA binding
- PMID: 10490612
- PMCID: PMC84664
- DOI: 10.1128/MCB.19.10.6729
Regulation of cell cycle transcription factor Swi4 through auto-inhibition of DNA binding
Abstract
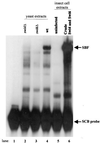
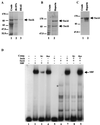
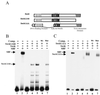
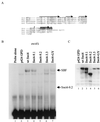
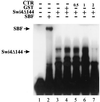
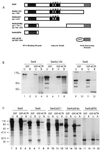
In Saccharomyces cerevisiae, two transcription factors, SBF (SCB binding factor) and MBF (MCB binding factor), promote the induction of gene expression at the G(1)/S-phase transition of the mitotic cell cycle. Swi4 and Mbp1 are the DNA binding components of SBF and MBF, respectively. The Swi6 protein is a common subunit of both transcription factors and is presumed to play a regulatory role. SBF binding to its target sequences, the SCBs, is a highly regulated event and requires the association of Swi4 with Swi6 through their C-terminal domains. Swi4 binding to SCBs is restricted to the late M and G(1) phases, when Swi6 is localized to the nucleus. We show that in contrast to Swi6, Swi4 remains nuclear throughout the cell cycle. This finding suggests that the DNA binding domain of Swi4 is inaccessible in the full-length protein when not complexed with Swi6. To explore this hypothesis, we expressed Swi4 and Swi6 in insect cells by using the baculovirus system. We determined that partially purified Swi4 cannot bind SCBs in the absence of Swi6. However, Swi4 derivatives carrying point mutations or alterations in the extreme C terminus were able to bind DNA or activate transcription in the absence of Swi6, and the C terminus of Swi4 inhibited Swi4 derivatives from binding DNA in trans. Full-length Swi4 was determined to be monomeric in solution, suggesting an intramolecular mechanism for auto-inhibition of binding to DNA by Swi4. We detected a direct in vitro interaction between a C-terminal fragment of Swi4 and the N-terminal 197 amino acids of Swi4, which contain the DNA binding domain. Together, our data suggest that intramolecular interactions involving the C-terminal region of Swi4 physically prevent the DNA binding domain from binding SCBs. The interaction of the carboxy-terminal region of Swi4 with Swi6 alleviates this inhibition, allowing Swi4 to bind DNA.
Figures



References
-
- Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. - PubMed
-
- Andrews B J, Herskowitz I. Identification of a DNA binding factor involved in cell-cycle control of the yeast HO gene. Cell. 1989;57:21–29. - PubMed
-
- Andrews B J, Herskowitz I. The yeast SWI4 protein contains a motif present in developmental regulators and is part of a complex involved in cell-cycle-dependent transcription. Nature. 1989;342:830–833. - PubMed
-
- Bassuk A G, Leiden J M. The role of Ets transcription factors in the development and function of the mammalian immune system. Adv Immunol. 1997;64:65–104. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
