Erf2, a novel gene product that affects the localization and palmitoylation of Ras2 in Saccharomyces cerevisiae
- PMID: 10490616
- PMCID: PMC84674
- DOI: 10.1128/MCB.19.10.6775
Erf2, a novel gene product that affects the localization and palmitoylation of Ras2 in Saccharomyces cerevisiae
Abstract
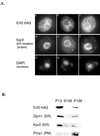
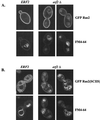
Plasma membrane localization of Ras requires posttranslational addition of farnesyl and palmitoyl lipid moieties to a C-terminal CaaX motif (C is cysteine, a is any aliphatic residue, X is the carboxy terminal residue). To better understand the relationship between posttranslational processing and the subcellular localization of Ras, a yeast genetic screen was undertaken based on the loss of function of a palmitoylation-dependent RAS2 allele. Mutations were identified in an uncharacterized open reading frame (YLR246w) that we have designated ERF2 and a previously described suppressor of hyperactive Ras, SHR5. ERF2 encodes a 41-kDa protein with four predicted transmembrane (TM) segments and a motif consisting of the amino acids Asp-His-His-Cys (DHHC) within a cysteine-rich domain (CRD), called DHHC-CRD. Mutations within the DHHC-CRD abolish Erf2 function. Subcellular fractionation and immunolocalization experiments reveal that Erf2 tagged with a triply iterated hemagglutinin epitope is an integral membrane protein that colocalizes with the yeast endoplasmic reticulum marker Kar2. Strains lacking ERF2 are viable, but they have a synthetic growth defect in the absence of RAS2 and partially suppress the heat shock sensitivity resulting from expression of the hyperactive RAS2(V19) allele. Ras2 proteins expressed in an erf2Delta strain have a reduced level of palmitoylation and are partially mislocalized to the vacuole. Based on these observations, we propose that Erf2 is a component of a previously uncharacterized Ras subcellular localization pathway. Putative members of an Erf2 family of proteins have been uncovered in yeast, plant, worm, insect, and mammalian genome databases, suggesting that Erf2 plays a role in Ras localization in all eucaryotes.
Figures





References
-
- Berger M, Schmidt M F G. Cell-free fatty acid acylation of Semliki forest viral polypeptides with microsomal membranes from eucaryotic cells. J Biol Chem. 1984;259:7245–7252. - PubMed
-
- Berthiaume L, Resh M D. Biochemical characterization of a palmitoyl acyltransferase activity that palmitoylates myristoylated proteins. J Biol Chem. 1995;270:22399–22405. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
