An autoregulatory loop controlling CYP1A1 gene expression: role of H(2)O(2) and NFI
- PMID: 10490621
- PMCID: PMC84679
- DOI: 10.1128/MCB.19.10.6825
An autoregulatory loop controlling CYP1A1 gene expression: role of H(2)O(2) and NFI
Abstract
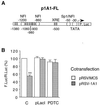
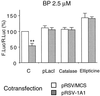
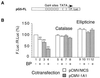
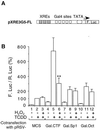
Cytochrome P450 1A1 (CYP1A1), like many monooxygenases, can produce reactive oxygen species during its catalytic cycle. Apart from the well-characterized xenobiotic-elicited induction, the regulatory mechanisms involved in the control of the steady-state activity of CYP1A1 have not been elucidated. We show here that reactive oxygen species generated from the activity of CYP1A1 limit the levels of induced CYP1A1 mRNAs. The mechanism involves the repression of the CYP1A1 gene promoter activity in a negative-feedback autoregulatory loop. Indeed, increasing the CYP1A1 activity by transfecting CYP1A1 expression vectors into hepatoma cells elicited an oxidative stress and led to the repression of a reporter gene driven by the CYP1A1 gene promoter. This negative autoregulation is abolished by ellipticine (an inhibitor of CYP1A1) and by catalase (which catalyzes H(2)O(2) catabolism), thus implying that H(2)O(2) is an intermediate. Down-regulation is also abolished by the mutation of the proximal nuclear factor I (NFI) site in the promoter. The transactivating domain of NFI/CTF was found to act in synergy with the arylhydrocarbon receptor pathway during the induction of CYP1A1 by 2,3,7,8-tetrachloro-p-dibenzodioxin. Using an NFI/CTF-Gal4 fusion, we show that NFI/CTF transactivating function is decreased by a high activity of CYP1A1. This regulation is also abolished by catalase or ellipticine. Consistently, the transactivating function of NFI/CTF is repressed in cells treated with H(2)O(2), a novel finding indicating that the transactivating domain of a transcription factor can be targeted by oxidative stress. In conclusion, an autoregulatory loop leads to the fine tuning of the CYP1A1 gene expression through the down-regulation of NFI activity by CYP1A1-based H(2)O(2) production. This mechanism allows a limitation of the potentially toxic CYP1A1 activity within the cell.
Figures
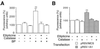
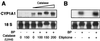

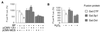

References
-
- Abdel-Razzak Z, Corcos L, Fautrel A, Campion J-P, Guillouzo A. Transforming growth factor-β1 down-regulates basal and polycyclic aromatic hydrocarbon-induced cytochromes P-450 1A1 and 1A2 in adult human hepatocytes in primary culture. Mol Pharmacol. 1994;46:1100–1110. - PubMed
-
- Alevizopoulos A, Dusserre Y, Tsai-Pflugfelder M, von der Weid T, Wahli W, Mermod N. A proline-rich TGF-β-responsive transcriptional activator interacts with histone H3. Genes Dev. 1995;9:3051–3066. - PubMed
-
- Barker C W, Fagan J B, Pasco D S. Down-regulation of P450 1A1 and 1A2 mRNA expression in isolated hepatocytes by oxidative stress. J Biol Chem. 1994;269:3985–3990. - PubMed
-
- Barker C W, Fagan J B, Pasco D S. Interleukin-1β suppresses the induction of P4501A1 and P4501A2 mRNAs in isolated hepatocytes. J Biol Chem. 1992;267:8050–8055. - PubMed
-
- Bondy S C, Naderi S. Contribution of hepatic cytochrome P450 systems to the generation of reactive oxygen species. Biochem Pharmacol. 1994;48:155–159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
