The essential functions of human Rad51 are independent of ATP hydrolysis
- PMID: 10490626
- PMCID: PMC84684
- DOI: 10.1128/MCB.19.10.6891
The essential functions of human Rad51 are independent of ATP hydrolysis
Abstract
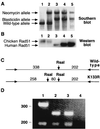
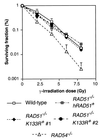
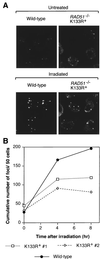
Genetic recombination and the repair of double-strand DNA breaks in Saccharomyces cerevisiae require Rad51, a homologue of the Escherichia coli RecA protein. In vitro, Rad51 binds DNA to form an extended nucleoprotein filament and catalyzes the ATP-dependent exchange of DNA between molecules with homologous sequences. Vertebrate Rad51 is essential for cell proliferation. Using site-directed mutagenesis of highly conserved residues of human Rad51 (hRad51) and gene targeting of the RAD51 locus in chicken DT40 cells, we examined the importance of Rad51's highly conserved ATP-binding domain. Mutant hRad51 incapable of ATP hydrolysis (hRad51K-133R) binds DNA less efficiently than the wild type but catalyzes strand exchange between homologous DNAs. hRad51 does not need to hydrolyze ATP to allow vertebrate cell proliferation, form nuclear foci, or repair radiation-induced DNA damage. However, cells expressing hRad51K-133R show greatly reduced targeted integration frequencies. These findings show that ATP hydrolysis is involved in DNA binding by hRad51 and suggest that the extent of DNA complexed with hRad51 in nucleoprotein influences the efficiency of recombination.
Figures
References
-
- Baumann P, Benson F E, Hajibagheri N, West S C. Purification of human Rad51 protein by selective spermidine precipitation. Mutat Res. 1997;384:65–72. - PubMed
-
- Baumann P, Benson F E, West S C. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. - PubMed
-
- Baumann P, West S C. Role of the human Rad51 protein in homologous recombination and double-stranded break repair. Trends Biochem Sci. 1998;23:247–251. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
