Delayed translational silencing of ceruloplasmin transcript in gamma interferon-activated U937 monocytic cells: role of the 3' untranslated region
- PMID: 10490627
- PMCID: PMC84685
- DOI: 10.1128/MCB.19.10.6898
Delayed translational silencing of ceruloplasmin transcript in gamma interferon-activated U937 monocytic cells: role of the 3' untranslated region
Abstract
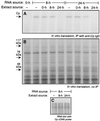
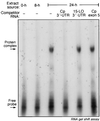
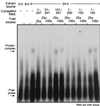
Ceruloplasmin (Cp) is an acute-phase protein with ferroxidase, amine oxidase, and pro- and antioxidant activities. The primary site of Cp synthesis in human adults is the liver, but it is also synthesized by cells of monocytic origin. We have shown that gamma interferon (IFN-gamma) induces the synthesis of Cp mRNA and protein in monocytic cells. We now report that the induced synthesis of Cp is terminated by a mechanism involving transcript-specific translational repression. Cp protein synthesis in U937 cells ceased after 16 h even in the presence of abundant Cp mRNA. RNA isolated from cells treated with IFN-gamma for 24 h exhibited a high in vitro translation rate, suggesting that the transcript was not defective. Ribosomal association of Cp mRNA was examined by sucrose centrifugation. When Cp synthesis was high, i.e., after 8 h of IFN-gamma treatment, Cp mRNA was primarily associated with polyribosomes. However, after 24 h, when Cp synthesis was low, Cp mRNA was primarily in the nonpolyribosomal fraction. Cytosolic extracts from cells treated with IFN-gamma for 24 h, but not for 8 h, contained a factor which blocked in vitro Cp translation. Inhibitor expression was cell type specific and present in extracts of human cells of myeloid origin, but not in several nonmyeloid cells. The inhibitory factor bound to the 3' untranslated region (3'-UTR) of Cp mRNA, as shown by restoration of in vitro translation by synthetic 3'-UTR added as a "decoy" and detection of a binding complex by RNA gel shift analysis. Deletion mapping of the Cp 3'-UTR indicated an internal 100-nucleotide region of the Cp 3'-UTR that was required for complex formation as well as for silencing of translation. Although transcript-specific translational control is common during development and differentiation and global translational control occurs during responses to cytokines and stress, to our knowledge, this is the first report of translational silencing of a specific transcript following cytokine activation.
Figures






References
-
- Askwith C, Eide D, Van Ho A, Bernard P S, Li L, Davis-Kaplan S, Sipe D M, Kaplan J. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell. 1994;76:403–410. - PubMed
-
- Attieh Z K, Mukhopadhyay C K, Seshadri V, Tripoulas N A, Fox P L. Ceruloplasmin ferroxidase activity stimulates cellular iron uptake by a trivalent cation-specific transport mechanism. J Biol Chem. 1999;274:1116–1123. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
