Sperm chromatin decondensation by template activating factor I through direct interaction with basic proteins
- PMID: 10490631
- PMCID: PMC84689
- DOI: 10.1128/MCB.19.10.6940
Sperm chromatin decondensation by template activating factor I through direct interaction with basic proteins
Abstract
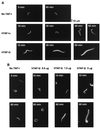
Template activating factor I (TAF-I) was originally identified as a host factor required for DNA replication and transcription of adenovirus genome complexed with viral basic proteins. Purified TAF-I was shown to bind to core histones and stimulate transcription from nucleosomal templates. Human TAF-I consists of two acidic proteins, TAF-Ialpha and TAF-Ibeta, which differ from each other only in their amino-terminal regions. Here, we report that TAF-I decondenses demembraned Xenopus sperm chromatin. Human TAF-Ibeta has a chromatin decondensation activity comparable to that of NAP-I, another histone binding protein, whereas TAF-Ialpha has only a weak activity. Analysis of molecular mechanisms underlying the chromatin decondensation by TAF-I revealed that TAF-I interacts directly with sperm basic proteins. Deletion of the TAF-I carboxyl-terminal acidic region abolishes the decondensation activity. Interestingly, the acidic region itself is not sufficient for decondensation, since an amino acid substitution mutant in the dimerization domain of TAF-I which has the intact acidic region does not support chromatin decondensation. We detected the beta form of TAF-I in Xenopus oocytes and eggs by immunoblotting, and the cloning of its cDNA led us to conclude that Xenopus TAF-Ibeta also decondenses sperm chromatin. These results suggest that TAF-I plays a role in remodeling higher-order chromatin structure as well as nucleosomal structure through direct interaction with chromatin basic proteins.
Figures



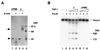




Similar articles
-
Coiled-coil structure-mediated dimerization of template activating factor-I is critical for its chromatin remodeling activity.J Mol Biol. 1999 Jul 9;290(2):547-57. doi: 10.1006/jmbi.1999.2898. J Mol Biol. 1999. PMID: 10390352
-
Histone- and chromatin-binding activity of template activating factor-I.FEBS Lett. 1999 Dec 17;463(3):285-8. doi: 10.1016/s0014-5793(99)01632-4. FEBS Lett. 1999. PMID: 10606739
-
Identification of distinct SET/TAF-Ibeta domains required for core histone binding and quantitative characterisation of the interaction.BMC Biochem. 2009 Apr 9;10:10. doi: 10.1186/1471-2091-10-10. BMC Biochem. 2009. PMID: 19358706 Free PMC article.
-
The role of nucleoplasmin in chromatin assembly and disassembly.Philos Trans R Soc Lond B Biol Sci. 1993 Mar 29;339(1289):263-9; discussion 268-9. doi: 10.1098/rstb.1993.0024. Philos Trans R Soc Lond B Biol Sci. 1993. PMID: 8098530 Review.
-
Histone chaperones and nucleosome assembly.Curr Opin Struct Biol. 2003 Feb;13(1):6-14. doi: 10.1016/s0959-440x(03)00002-2. Curr Opin Struct Biol. 2003. PMID: 12581654 Review.
Cited by
-
SET protein interacts with intracellular domains of the gonadotropin-releasing hormone receptor and differentially regulates receptor signaling to cAMP and calcium in gonadotrope cells.J Biol Chem. 2013 Jan 25;288(4):2641-54. doi: 10.1074/jbc.M112.388876. Epub 2012 Dec 11. J Biol Chem. 2013. PMID: 23233674 Free PMC article.
-
Eviction of linker histone H1 by NAP-family histone chaperones enhances activated transcription.Epigenetics Chromatin. 2015 Sep 4;8:30. doi: 10.1186/s13072-015-0022-8. eCollection 2015. Epigenetics Chromatin. 2015. PMID: 26339295 Free PMC article.
-
Function of homo- and hetero-oligomers of human nucleoplasmin/nucleophosmin family proteins NPM1, NPM2 and NPM3 during sperm chromatin remodeling.Nucleic Acids Res. 2012 Jun;40(11):4861-78. doi: 10.1093/nar/gks162. Epub 2012 Feb 22. Nucleic Acids Res. 2012. PMID: 22362753 Free PMC article.
-
Yeast Nap1-binding protein Nbp2p is required for mitotic growth at high temperatures and for cell wall integrity.Genetics. 2003 Oct;165(2):517-29. doi: 10.1093/genetics/165.2.517. Genetics. 2003. PMID: 14573466 Free PMC article.
-
PP2A complex disruptor SET prompts widespread hypertranscription of growth-essential genes in the pancreatic cancer cells.Sci Adv. 2024 Jan 26;10(4):eadk6633. doi: 10.1126/sciadv.adk6633. Epub 2024 Jan 26. Sci Adv. 2024. PMID: 38277454 Free PMC article.
References
-
- Adachi Y, Pavlakis G N, Copeland T D. Identification and characterization of SET, a nuclear phosphoprotein encoded by the translocation break point in acute undifferentiated leukemia. J Biol Chem. 1994;269:2258–2262. - PubMed
-
- Adler H T, Nallaseth F S, Walter G, Tkachuk D C. HRX leukemic fusion proteins form a heterocomplex with the leukemia-associated protein SET and protein phosphatase 2A. J Biol Chem. 1997;272:28407–28414. - PubMed
-
- Almouzni G, Wolffe A P. Nuclear assembly, structure, and function: the use of Xenopus in vitro systems. Exp Cell Res. 1993;205:1–15. - PubMed
-
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
