Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae
- PMID: 10490634
- PMCID: PMC84692
- DOI: 10.1128/MCB.19.10.6972
Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae
Abstract
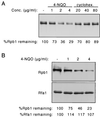
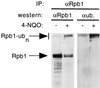
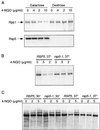
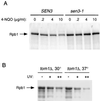
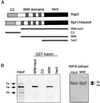
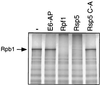
Rsp5 is an E3 ubiquitin-protein ligase of Saccharomyces cerevisiae that belongs to the hect domain family of E3 proteins. We have previously shown that Rsp5 binds and ubiquitinates the largest subunit of RNA polymerase II, Rpb1, in vitro. We show here that Rpb1 ubiquitination and degradation are induced in vivo by UV irradiation and by the UV-mimetic compound 4-nitroquinoline-1-oxide (4-NQO) and that a functional RSP5 gene product is required for this effect. The 26S proteasome is also required; a mutation of SEN3/RPN2 (sen3-1), which encodes an essential regulatory subunit of the 26S proteasome, partially blocks 4-NQO-induced degradation of Rpb1. These results suggest that Rsp5-mediated ubiquitination and degradation of Rpb1 are components of the response to DNA damage. A human WW domain-containing hect (WW-hect) E3 protein closely related to Rsp5, Rpf1/hNedd4, also binds and ubiquitinates both yeast and human Rpb1 in vitro, suggesting that Rpf1 and/or another WW-hect E3 protein mediates UV-induced degradation of the large subunit of polymerase II in human cells.
Figures


References
-
- Beaudenon, S. L., and J. M. Huibregtse. Unpublished results.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
