NRIF3 is a novel coactivator mediating functional specificity of nuclear hormone receptors
- PMID: 10490654
- PMCID: PMC84712
- DOI: 10.1128/MCB.19.10.7191
NRIF3 is a novel coactivator mediating functional specificity of nuclear hormone receptors
Abstract
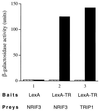
Many nuclear receptors are capable of recognizing similar DNA elements. The molecular event(s) underlying the functional specificities of these receptors (in regulating the expression of their native target genes) is a very important issue that remains poorly understood. Here we report the cloning and analysis of a novel nuclear receptor coactivator (designated NRIF3) that exhibits a distinct receptor specificity. Fluorescence microscopy shows that NRIF3 localizes to the cell nucleus. The yeast two-hybrid and/or in vitro binding assays indicated that NRIF3 specifically interacts with the thyroid hormone receptor (TR) and retinoid X receptor (RXR) in a ligand-dependent fashion but does not bind to the retinoic acid receptor, vitamin D receptor, progesterone receptor, glucocorticoid receptor, or estrogen receptor. Functional experiments showed that NRIF3 significantly potentiates TR- and RXR-mediated transactivation in vivo but has little effect on other examined nuclear receptors. Domain and mutagenesis analyses indicated that a novel C-terminal domain in NRIF3 plays an essential role in its specific interaction with liganded TR and RXR while the N-terminal LXXLL motif plays a minor role in allowing optimum interaction. Computer modeling and subsequent experimental analysis suggested that the C-terminal domain of NRIF3 directly mediates interaction with liganded receptors through an LXXIL (a variant of the canonical LXXLL) module while the other part of the NRIF3 protein may still play a role in conferring its receptor specificity. Identification of a coactivator with such a unique receptor specificity may provide new insight into the molecular mechanism(s) of receptor-mediated transcriptional activation as well as the functional specificities of nuclear receptors.
Figures



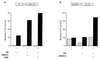





Similar articles
-
Domain structure of the NRIF3 family of coregulators suggests potential dual roles in transcriptional regulation.Mol Cell Biol. 2001 Dec;21(24):8371-84. doi: 10.1128/MCB.21.24.8371-8384.2001. Mol Cell Biol. 2001. PMID: 11713274 Free PMC article.
-
A regulatory role for RIP140 in nuclear receptor activation.Mol Endocrinol. 1998 Jun;12(6):864-81. doi: 10.1210/mend.12.6.0123. Mol Endocrinol. 1998. PMID: 9626662
-
Analysis of the functional role of steroid receptor coactivator-1 in ligand-induced transactivation by thyroid hormone receptor.Mol Endocrinol. 1997 Jun;11(6):755-67. doi: 10.1210/mend.11.6.0003. Mol Endocrinol. 1997. PMID: 9171239
-
Ligand-dependent interaction of nuclear receptors with potential transcriptional intermediary factors (mediators).Philos Trans R Soc Lond B Biol Sci. 1996 Apr 29;351(1339):569-78. doi: 10.1098/rstb.1996.0056. Philos Trans R Soc Lond B Biol Sci. 1996. PMID: 8735280 Review.
-
Mechanisms and significance of nuclear receptor auto- and cross-regulation.Gen Comp Endocrinol. 2011 Jan 1;170(1):3-17. doi: 10.1016/j.ygcen.2010.03.013. Epub 2010 Mar 23. Gen Comp Endocrinol. 2011. PMID: 20338175 Free PMC article. Review.
Cited by
-
Phosphorylation of CENP-R by Aurora B regulates kinetochore-microtubule attachment for accurate chromosome segregation.J Mol Cell Biol. 2022 Sep 27;14(7):mjac051. doi: 10.1093/jmcb/mjac051. J Mol Cell Biol. 2022. PMID: 36069839 Free PMC article.
-
Rational discovery of novel nuclear hormone receptor antagonists.Proc Natl Acad Sci U S A. 2000 Feb 1;97(3):1008-13. doi: 10.1073/pnas.97.3.1008. Proc Natl Acad Sci U S A. 2000. PMID: 10655475 Free PMC article.
-
In silico discovery of novel retinoic acid receptor agonist structures.BMC Struct Biol. 2001;1:1. doi: 10.1186/1472-6807-1-1. Epub 2001 Jun 4. BMC Struct Biol. 2001. PMID: 11405897 Free PMC article.
-
Peptides derived from the integrin β cytoplasmic tails inhibit angiogenesis.Cell Commun Signal. 2018 Jul 3;16(1):38. doi: 10.1186/s12964-018-0248-8. Cell Commun Signal. 2018. PMID: 29970081 Free PMC article.
-
High-throughput discovery and characterization of viral transcriptional effectors in human cells.Cell Syst. 2023 Jun 21;14(6):482-500.e8. doi: 10.1016/j.cels.2023.05.008. Cell Syst. 2023. PMID: 37348463 Free PMC article.
References
-
- Abagyan R A, Totrov M M, Kuznetsov D A. ICM: a new method for structure modeling and design: applications to docking and structure prediction from the distorted native conformation. J Comp Chem. 1994;15:488–506.
-
- Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. - PubMed
-
- Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. - PubMed
-
- Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
