De novo synthesis of sphingolipids is required for cell survival by down-regulating c-Jun N-terminal kinase in Drosophila imaginal discs
- PMID: 10490662
- PMCID: PMC84720
- DOI: 10.1128/MCB.19.10.7276
De novo synthesis of sphingolipids is required for cell survival by down-regulating c-Jun N-terminal kinase in Drosophila imaginal discs
Abstract
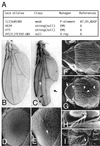
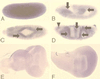
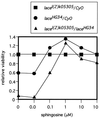
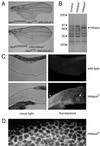
Mitogen-activated protein kinase (MAPK) is a conserved eukaryotic signaling factor that mediates various signals, cumulating in the activation of transcription factors. Extracellular signal-regulated kinase (ERK), a MAPK, is activated through phosphorylation by the kinase MAPK/ERK kinase (MEK). To elucidate the extent of the involvement of ERK in various aspects of animal development, we searched for a Drosophila mutant which responds to elevated MEK activity and herein identified a lace mutant. Mutants with mild lace alleles grow to become adults with multiple aberrant morphologies in the appendages, compound eye, and bristles. These aberrations were suppressed by elevated MEK activity. Structural and transgenic analyses of the lace cDNA have revealed that the lace gene product is a membrane protein similar to the yeast protein LCB2, a subunit of serine palmitoyltransferase (SPT), which catalyzes the first step of sphingolipid biosynthesis. In fact, SPT activity in the fly expressing epitope-tagged Lace was absorbed by epitope-specific antibody. The number of dead cells in various imaginal discs of a lace hypomorph was considerably increased, thereby ectopically activating c-Jun N-terminal kinase (JNK), another MAPK. These results account for the adult phenotypes of the lace mutant and suppression of the phenotypes by elevated MEK activity: we hypothesize that mutation of lace causes decreased de novo synthesis of sphingolipid metabolites, some of which are signaling molecules, and one or more of these changes activates JNK to elicit apoptosis. The ERK pathway may be antagonistic to the JNK pathway in the control of cell survival.
Figures





References
-
- Adachi-Yamada, T. Unpublished data.
-
- Adachi-Yamada T, Nakamura M, Irie K, Tomoyasu Y, Sano Y, Mori E, Goto S, Ueno N, Nishida Y, Matsumoto K. p38 mitogen-activated protein kinase can be involved in transforming growth factor β superfamily signal transduction in Drosophila wing morphogenesis. Mol Cell Biol. 1999;19:2322–2329. - PMC - PubMed
-
- Adachi-Yamada T, Fujimura-Kamada K, Nishida Y, Matsumoto K. Distortion of proximodistal information causes JNK-dependent apoptosis in Drosophila wing. Nature. 1999;400:166–169. - PubMed
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Ashburner M. Drosophila, a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
