Nuclear organization of mammalian genomes. Polar chromosome territories build up functionally distinct higher order compartments
- PMID: 10491386
- PMCID: PMC2156120
- DOI: 10.1083/jcb.146.6.1211
Nuclear organization of mammalian genomes. Polar chromosome territories build up functionally distinct higher order compartments
Abstract
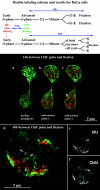
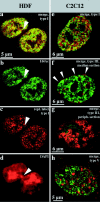
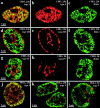
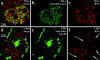
We investigated the nuclear higher order compartmentalization of chromatin according to its replication timing (Ferreira et al. 1997) and the relations of this compartmentalization to chromosome structure and the spatial organization of transcription. Our aim was to provide a comprehensive and integrated view on the relations between chromosome structure and functional nuclear architecture. Using different mammalian cell types, we show that distinct higher order compartments whose DNA displays a specific replication timing are stably maintained during all interphase stages. The organizational principle is clonally inherited. We directly demonstrate the presence of polar chromosome territories that align to build up higher order compartments, as previously suggested (Ferreira et al. 1997). Polar chromosome territories display a specific orientation of early and late replicating subregions that correspond to R- or G/C-bands of mitotic chromosomes. Higher order compartments containing G/C-bands replicating during the second half of the S phase display no transcriptional activity detectable by BrUTP pulse labeling and show no evidence of transcriptional competence. Transcriptionally competent and active chromatin is confined to a coherent compartment within the nuclear interior that comprises early replicating R-band sequences. As a whole, the data provide an integrated view on chromosome structure, nuclear higher order compartmentalization, and their relation to the spatial organization of functional nuclear processes.
Figures






References
-
- Andrulis E.D., Neimann A.M., Zappulla D.C., Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. - PubMed
-
- Belmont A.S., Bignone F., Ts'o P.O.P. The relative intranuclear positions of Barr bodies in XXX non-transformed human fibroblasts. Exp. Cell Res. 1986;165:165–179. - PubMed
-
- Berezney R., Mortillaro M.J., Ma H., Wei X., Samarabandu J. The nuclear matrixa structural milieu for genomic function. Int. Rev. Cytol. 1995;162A:1–65. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

