Plasmid RK2 ParB protein: purification and nuclease properties
- PMID: 10498713
- PMCID: PMC103628
- DOI: 10.1128/JB.181.19.6010-6018.1999
Plasmid RK2 ParB protein: purification and nuclease properties
Abstract
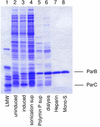
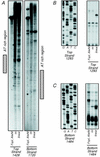
The parCBA operon of the 3.2-kb stabilization region of plasmid RK2 encodes three cotranslated proteins. ParA mediates site-specific recombination to resolve plasmid multimers, ParB has been shown to be a nuclease, and the function of ParC is unknown. In this study ParB was overexpressed by cotranslation with ParC in Escherichia coli by using a plasmid construct that contained the parC and parB genes under the control of the T7 promoter. Purification was achieved by treatment of extracts with Polymin P, followed by ammonium sulfate precipitation and heparin and ion-exchange chromatography. Sizing-column analysis indicated that ParB exists as a monomer in solution. Analysis of the enzymatic properties of purified ParB indicated that the protein preferentially cleaves single-stranded DNA. ParB also nicks supercoiled plasmid DNA preferably at sites with potential single-stranded character, like AT-rich regions and sequences that can form cruciform structures. ParB also exhibits 5'-->3' exonuclease activity. This ParB activity on a 5'-end-labeled, double-stranded DNA substrate produces a 3', 5'-phosphorylated dinucleotide which is further cleaved to a 3', 5'-phosphorylated mononucleotide. The role of the ParB endonuclease and exonuclease activities in plasmid RK2 stabilization remains to be determined.
Figures






Similar articles
-
Molecular analysis of the pRA2 partitioning region: ParB autoregulates parAB transcription and forms a nucleoprotein complex with the plasmid partition site, parS.Mol Microbiol. 2001 May;40(3):621-33. doi: 10.1046/j.1365-2958.2001.02405.x. Mol Microbiol. 2001. PMID: 11359568
-
A native cruciform DNA structure probed in bacteria by recombinant T7 endonuclease.J Biol Chem. 1987 Aug 15;262(23):11364-8. J Biol Chem. 1987. PMID: 3038915
-
Characterization of the stable maintenance properties of the par region of broad-host-range plasmid RK2.J Bacteriol. 1996 Apr;178(7):2086-93. doi: 10.1128/jb.178.7.2086-2093.1996. J Bacteriol. 1996. PMID: 8606188 Free PMC article.
-
Contribution of different segments of the par region to stable maintenance of the broad-host-range plasmid RK2.J Bacteriol. 1997 Oct;179(20):6472-9. doi: 10.1128/jb.179.20.6472-6479.1997. J Bacteriol. 1997. PMID: 9335298 Free PMC article.
-
Role of the parCBA operon of the broad-host-range plasmid RK2 in stable plasmid maintenance.J Bacteriol. 1998 Nov;180(22):6023-30. doi: 10.1128/JB.180.22.6023-6030.1998. J Bacteriol. 1998. PMID: 9811663 Free PMC article.
Cited by
-
Wolbachia prophage DNA adenine methyltransferase genes in different Drosophila-Wolbachia associations.PLoS One. 2011 May 6;6(5):e19708. doi: 10.1371/journal.pone.0019708. PLoS One. 2011. PMID: 21573076 Free PMC article.
-
TasA-tasB, a new putative toxin-antitoxin (TA) system from Bacillus thuringiensis pGI1 plasmid is a widely distributed composite mazE-doc TA system.BMC Genomics. 2006 Oct 13;7:259. doi: 10.1186/1471-2164-7-259. BMC Genomics. 2006. PMID: 17038198 Free PMC article.
-
Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics.Biol Direct. 2012 Jun 25;7:18. doi: 10.1186/1745-6150-7-18. Biol Direct. 2012. PMID: 22731697 Free PMC article.
-
New biochemistry in the Rhodanese-phosphatase superfamily: emerging roles in diverse metabolic processes, nucleic acid modifications, and biological conflicts.NAR Genom Bioinform. 2023 Mar 23;5(1):lqad029. doi: 10.1093/nargab/lqad029. eCollection 2023 Mar. NAR Genom Bioinform. 2023. PMID: 36968430 Free PMC article.
-
Introducing Casbah, Kronus, and MmasiCarm, Members of the Mycobacteriophage Subcluster B3.Phage (New Rochelle). 2024 Jun 21;5(2):84-90. doi: 10.1089/phage.2024.0004. eCollection 2024 Jun. Phage (New Rochelle). 2024. PMID: 39119203 Free PMC article.
References
-
- Adams D E, Shekhtman E M, Zechiedrich E L, Schmid M B, Cozzarelli N R. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell. 1992;71:277–288. - PubMed
-
- Alvarado-Urbina G, Sathe G M, Liu W C, Gillen M F, Duck P D, Bender R, Ogilvie K K. Automated synthesis of gene fragments. Science. 1981;214:270–274. - PubMed
-
- Austin S, Abeles A. Partition of unit-copy miniplasmids to daughter cells. II. The partition region of miniplasmid P1 encodes an essential protein and a centromere-like site at which it acts. J Mol Biol. 1983;169:373–387. - PubMed
-
- Bernard P, Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-Topoisomerase II complexes. J Mol Biol. 1992;226:735–745. - PubMed
-
- Bravo A, de Torrontegui G, Diaz R. Identification of components of a new stability system of R1, ParD, that is close to the origin of replication of the plasmid. Mol Gen Genet. 1987;210:101–110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

