Use of heme-protein complexes by the Yersinia enterocolitica HemR receptor: histidine residues are essential for receptor function
- PMID: 10498719
- PMCID: PMC103634
- DOI: 10.1128/JB.181.19.6063-6072.1999
Use of heme-protein complexes by the Yersinia enterocolitica HemR receptor: histidine residues are essential for receptor function
Abstract
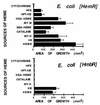
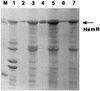
The abilities of two bacterial active heme transporters, HmbR of Neisseria meningitidis and HemR of Yersinia enterocolitica, to use different heme sources were compared. While HmbR-expressing cells used only hemoglobin (Hb) and heme, HemR-expressing bacteria were able to grow on Hb, heme, myoglobin, hemopexin, catalase, human and bovine serum albumin-heme, and haptoglobin-hemoglobin complexes as sources of iron. Expression of functional HemR allowed Escherichia coli cells to respond to heme-containing peptides, microperoxidases MP-8, MP-9, and MP-11, suggesting the ability of HemR to transport heme covalently linked to other molecules. Comparison of HemR with other heme receptors identified several highly conserved histidine residues as well as two conserved amino acid motifs, the FRAP and NPNL boxes. A site-directed mutagenesis approach was used to investigate the roles of His128, His192, His352, and His461 residues in HemR function. The HemR receptor with histidine changed to lysine at position 128 (HemR(H128K)), HemR(H461L), HemR(H461A), and HemR(H128A,H461A) mutant receptors were unable to use Hb, human serum albumin-heme, and myoglobin as sources of porphyrin and iron. Utilization of free heme was also severely affected, with some residual heme uptake in cells expressing HemR(H128K), HemR(H461A), and HemR(H461L). Conversely, the HemR(H192T), HemR(H352A), HemR(H352K), and HemR(H192T,H352K) mutant receptors were fully functional. All mutant HemR proteins were expressed in the outer membrane at levels similar to that of the wild-type HemR receptor. Nonfunctional HemRs were able to bind heme- and Hb-agarose. A hypothetical model of the HemR function in which two conserved histidine residues, His128 and His461, participate in the transport of heme through the receptor pore is postulated.
Figures







Similar articles
-
Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria.EMBO J. 1992 Dec;11(12):4359-67. doi: 10.1002/j.1460-2075.1992.tb05535.x. EMBO J. 1992. PMID: 1425573 Free PMC article.
-
OmpR-Mediated Transcriptional Regulation and Function of Two Heme Receptor Proteins of Yersinia enterocolitica Bio-Serotype 2/O:9.Front Cell Infect Microbiol. 2018 Sep 20;8:333. doi: 10.3389/fcimb.2018.00333. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30294593 Free PMC article.
-
Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis.J Bacteriol. 2000 Oct;182(20):5737-48. doi: 10.1128/JB.182.20.5737-5748.2000. J Bacteriol. 2000. PMID: 11004172 Free PMC article.
-
Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores.Curr Opin Microbiol. 2000 Apr;3(2):215-20. doi: 10.1016/s1369-5274(00)00078-3. Curr Opin Microbiol. 2000. PMID: 10744995 Review.
-
The physical chemistry of bacterial outer-membrane siderophore receptor proteins.Met Ions Biol Syst. 1998;35:355-401. Met Ions Biol Syst. 1998. PMID: 9444764 Review. No abstract available.
Cited by
-
HasB, the Serratia marcescens TonB paralog, is specific to HasR.J Bacteriol. 2008 Jan;190(1):21-7. doi: 10.1128/JB.01389-07. Epub 2007 Oct 19. J Bacteriol. 2008. PMID: 17951376 Free PMC article.
-
BhuR, a virulence-associated outer membrane protein of Bordetella avium, is required for the acquisition of iron from heme and hemoproteins.Infect Immun. 2002 Oct;70(10):5390-403. doi: 10.1128/IAI.70.10.5390-5403.2002. Infect Immun. 2002. PMID: 12228263 Free PMC article.
-
The Bordetella bhu locus is required for heme iron utilization.J Bacteriol. 2001 Jul;183(14):4278-87. doi: 10.1128/JB.183.14.4278-4287.2001. J Bacteriol. 2001. PMID: 11418569 Free PMC article.
-
Identification of a hemin utilization protein of Moraxella catarrhalis (HumA).Infect Immun. 2004 Nov;72(11):6426-32. doi: 10.1128/IAI.72.11.6426-6432.2004. Infect Immun. 2004. PMID: 15501773 Free PMC article.
-
An Exposed Outer Membrane Hemin-Binding Protein Facilitates Hemin Transport by a TonB-Dependent Receptor in Riemerella anatipestifer.Appl Environ Microbiol. 2021 Jul 13;87(15):e0036721. doi: 10.1128/AEM.00367-21. Epub 2021 Jul 13. Appl Environ Microbiol. 2021. PMID: 33990314 Free PMC article.
References
-
- Aisen P, Leibman A, Zweier J. Stoichiometric and site characteristics of the binding of iron to human transferrin. J Biol Chem. 1978;253:1930–1937. - PubMed
-
- Armstrong S K, Francis C L, McIntosh M A. Molecular analysis of the Escherichia coli ferric enterobactin receptor FepA. J Biol Chem. 1990;265:14536–14543. - PubMed
-
- Beale S I. Biosynthesis of hemes. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 731–748.
-
- Buchanan S K, Smith B S, Vankatramani L, Xia D, Esser L, Palnitkar M, Chakraborty R, Van der Helm D, Deisenhofer J. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat Struct Biol. 1999;6:56–63. - PubMed
-
- Bunn H F, Jandl J H. Exchange of heme among hemoglobins and between globin and albumin. J Biol Chem. 1968;243:465–475. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

