Phylogenetic analysis of L4-mediated autogenous control of the S10 ribosomal protein operon
- PMID: 10498727
- PMCID: PMC103642
- DOI: 10.1128/JB.181.19.6124-6132.1999
Phylogenetic analysis of L4-mediated autogenous control of the S10 ribosomal protein operon
Abstract
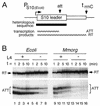
We investigated the regulation of the S10 ribosomal protein (r-protein) operon among members of the gamma subdivision of the proteobacteria, which includes Escherichia coli. In E. coli, this 11-gene operon is autogenously controlled by r-protein L4. This regulation requires specific determinants within the untranslated leader of the mRNA. Secondary structure analysis of the S10 leaders of five enterobacteria (Salmonella typhimurium, Citrobacter freundii, Yersinia enterocolitica, Serratia marcescens, and Morganella morganii) and two nonenteric members of the gamma subdivision (Haemophilus influenzae and Vibrio cholerae) shows that these foreign leaders share significant structural homology with the E. coli leader, particularly in the region which is critical for L4-mediated autogenous control in E. coli. Moreover, these heterologous leaders produce a regulatory response to L4 oversynthesis in E. coli. Our results suggest that an E. coli-like L4-mediated regulatory mechanism may operate in all of these species. However, the mechanism is not universally conserved among the gamma subdivision members, since at least one, Pseudomonas aeruginosa, does not contain the required S10 leader features, and its leader cannot provide the signals for regulation by L4 in E. coli. We speculate that L4-mediated autogenous control developed during the evolution of the gamma branch of proteobacteria.
Figures



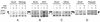



References
-
- Beck C, Ingraham J, Maaløe O, Neuhard J. Relationship between the concentration of nucleoside triphosphates and the rate of synthesis of RNA. J Mol Biol. 1973;78:117–121. - PubMed
-
- Clark D J, Maaløe O. DNA replication and the division cycle in Escherichia coli. J Mol Biol. 1967;23:99–112.
-
- Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

