FlbT couples flagellum assembly to gene expression in Caulobacter crescentus
- PMID: 10498731
- PMCID: PMC103646
- DOI: 10.1128/JB.181.19.6160-6170.1999
FlbT couples flagellum assembly to gene expression in Caulobacter crescentus
Abstract
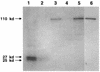
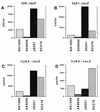
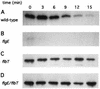
The biogenesis of the polar flagellum of Caulobacter crescentus is regulated by the cell cycle as well as by a trans-acting regulatory hierarchy that functions to couple flagellum assembly to gene expression. The assembly of early flagellar structures (MS ring, switch, and flagellum-specific secretory system) is required for the transcription of class III genes, which encode the remainder of the basal body and the external hook structure. Similarly, the assembly of class III gene-encoded structures is required for the expression of the class IV flagellins, which are incorporated into the flagellar filament. Here, we demonstrate that mutations in flbT, a flagellar gene of unknown function, can restore flagellin protein synthesis and the expression of fljK::lacZ (25-kDa flagellin) protein fusions in class III flagellar mutants. These results suggest that FlbT functions to negatively regulate flagellin expression in the absence of flagellum assembly. Deletion analysis shows that sequences within the 5' untranslated region of the fljK transcript are sufficient for FlbT regulation. To determine the mechanism of FlbT-mediated regulation, we assayed the stability of fljK mRNA. The half-life (t(1/2)) of fljK mRNA in wild-type cells was approximately 11 min and was reduced to less than 1.5 min in a flgE (hook) mutant. A flgE flbT double mutant exhibited an mRNA t(1/2) of greater than 30 min. This suggests that the primary effect of FlbT regulation is an increased turnover of flagellin mRNA. The increased t(1/2) of fljK mRNA in a flbT mutant has consequences for the temporal expression of fljK. In contrast to the case for wild-type cells, fljK::lacZ protein fusions in the mutant are expressed almost continuously throughout the C. crescentus cell cycle, suggesting that coupling of flagellin gene expression to assembly has a critical influence on regulating cell cycle expression.
Figures
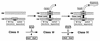


Similar articles
-
FlbT, the post-transcriptional regulator of flagellin synthesis in Caulobacter crescentus, interacts with the 5' untranslated region of flagellin mRNA.Mol Microbiol. 2000 Oct;38(1):41-52. doi: 10.1046/j.1365-2958.2000.02108.x. Mol Microbiol. 2000. PMID: 11029689
-
Posttranscriptional regulation of Caulobacter flagellin genes by a late flagellum assembly checkpoint.J Bacteriol. 1997 Apr;179(7):2281-8. doi: 10.1128/jb.179.7.2281-2288.1997. J Bacteriol. 1997. PMID: 9079914 Free PMC article.
-
The conserved flaF gene has a critical role in coupling flagellin translation and assembly in Caulobacter crescentus.Mol Microbiol. 2005 Aug;57(4):1127-42. doi: 10.1111/j.1365-2958.2005.04745.x. Mol Microbiol. 2005. PMID: 16091049
-
Regulation of flagellar assembly.Curr Opin Microbiol. 2002 Apr;5(2):160-5. doi: 10.1016/s1369-5274(02)00302-8. Curr Opin Microbiol. 2002. PMID: 11934612 Review.
-
Regulation of the Caulobacter flagellar gene hierarchy; not just for motility.Mol Microbiol. 1997 Apr;24(2):233-9. doi: 10.1046/j.1365-2958.1997.3281691.x. Mol Microbiol. 1997. PMID: 9159510 Review.
Cited by
-
Global regulation of gene expression and cell differentiation in Caulobacter crescentus in response to nutrient availability.J Bacteriol. 2010 Feb;192(3):819-33. doi: 10.1128/JB.01240-09. Epub 2009 Nov 30. J Bacteriol. 2010. PMID: 19948804 Free PMC article.
-
Multiple Flagellin Proteins Have Distinct and Synergistic Roles in Agrobacterium tumefaciens Motility.J Bacteriol. 2018 Nov 6;200(23):e00327-18. doi: 10.1128/JB.00327-18. Print 2018 Dec 1. J Bacteriol. 2018. PMID: 30201783 Free PMC article.
-
Transcriptional Control of the Lateral-Flagellar Genes of Bradyrhizobium diazoefficiens.J Bacteriol. 2017 Jul 11;199(15):e00253-17. doi: 10.1128/JB.00253-17. Print 2017 Aug 1. J Bacteriol. 2017. PMID: 28533217 Free PMC article.
-
Flagellar Structures from the Bacterium Caulobacter crescentus and Implications for Phage ϕ CbK Predation of Multiflagellin Bacteria.J Bacteriol. 2021 Feb 8;203(5):e00399-20. doi: 10.1128/JB.00399-20. Print 2021 Feb 8. J Bacteriol. 2021. PMID: 33288623 Free PMC article.
-
A family of six flagellin genes contributes to the Caulobacter crescentus flagellar filament.J Bacteriol. 2000 Sep;182(17):5001-4. doi: 10.1128/JB.182.17.5001-5004.2000. J Bacteriol. 2000. PMID: 10940048 Free PMC article.
References
-
- Alley M R K, Maddock J, Shapiro L. Requirement of the carboxyl terminus of a bacterial chemoreceptor for its targeted proteolysis. Science. 1993;259:1754–1757. - PubMed
-
- Anderson D K, Ohta N, Wu J, Newton A. Regulation of the Caulobacter crescentus rpoN gene and function of the purified sigma 54 in flagellar gene transcription. Mol Gen Genet. 1995;246:697–706. - PubMed
-
- Anderson, P. E., and J. W. Gober. Unpublished data.
-
- Ausubel F M, Brent R, Kingston R E, Moore D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1989.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

