The structure of the human betaII-tryptase tetramer: fo(u)r better or worse
- PMID: 10500112
- PMCID: PMC34230
- DOI: 10.1073/pnas.96.20.10984
The structure of the human betaII-tryptase tetramer: fo(u)r better or worse
Abstract
Tryptases, the predominant serine proteinases of human mast cells, have recently been implicated as mediators in the pathogenesis of allergic and inflammatory conditions, most notably asthma. Their distinguishing features, their activity as a heparin-stabilized tetramer and resistance to most proteinaceous inhibitors, are perfectly explained by the 3-A crystal structure of human betaII-tryptase in complex with 4-amidinophenylpyruvic acid. The tetramer consists of four quasiequivalent monomers arranged in a flat frame-like structure. The active centers are directed toward a central pore whose narrow openings of approximately 40 A x 15 A govern the interaction with macromolecular substrates and inhibitors. The tryptase monomer exhibits the overall fold of trypsin-like serine proteinases but differs considerably in the conformation of six surface loops arranged around the active site. These loops border and shape the active site cleft to a large extent and form all contacts with neighboring monomers via two distinct interfaces. The smaller of these interfaces, which is exclusively hydrophobic, can be stabilized by the binding of heparin chains to elongated patches of positively charged residues on adjacent monomers or, alternatively, by high salt concentrations in vitro. On tetramer dissociation, the monomers are likely to undergo transformation into a zymogen-like conformation that is favored and stabilized by intramonomer interactions. The structure thus provides an improved understanding of the unique properties of the biologically active tryptase tetramer in solution and will be an incentive for the rational design of mono- and multifunctional tryptase inhibitors.
Figures



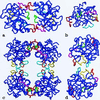




References
-
- Pallaoro M, Fejzo M S, Shayesteh L, Blount J L, Caughey G H. J Biol Chem. 1999;274:3355–3362. - PubMed
-
- Schwartz L B, Irani A M, Roller K, Castells M C, Schechter N M. J Immunol. 1987;138:2611–2615. - PubMed
-
- Xia H Z, Kepley C L, Sakai K, Chelliah J, Irani A M, Schwartz L B. J Immunol. 1995;154:5472–5480. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

