A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood
- PMID: 10500120
- PMCID: PMC34238
- DOI: 10.1073/pnas.96.20.11041
A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood
Abstract
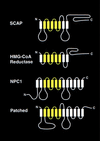
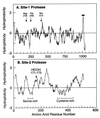
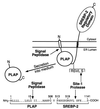
The integrity of cell membranes is maintained by a balance between the amount of cholesterol and the amounts of unsaturated and saturated fatty acids in phospholipids. This balance is maintained by membrane-bound transcription factors called sterol regulatory element-binding proteins (SREBPs) that activate genes encoding enzymes of cholesterol and fatty acid biosynthesis. To enhance transcription, the active NH(2)-terminal domains of SREBPs are released from endoplasmic reticulum membranes by two sequential cleavages. The first is catalyzed by Site-1 protease (S1P), a membrane-bound subtilisin-related serine protease that cleaves the hydrophilic loop of SREBP that projects into the endoplasmic reticulum lumen. The second cleavage, at Site-2, requires the action of S2P, a hydrophobic protein that appears to be a zinc metalloprotease. This cleavage is unusual because it occurs within a membrane-spanning domain of SREBP. Sterols block SREBP processing by inhibiting S1P. This response is mediated by SREBP cleavage-activating protein (SCAP), a regulatory protein that activates S1P and also serves as a sterol sensor, losing its activity when sterols overaccumulate in cells. These regulated proteolytic cleavage reactions are ultimately responsible for controlling the level of cholesterol in membranes, cells, and blood.
Figures


References
-
- Devaux P F. Curr Opin Struct Biol. 1993;3:489–494.
-
- Simons K, Ikonen E. Nature (London) 1997;387:569–572. - PubMed
-
- Anderson R G W. Annu Rev Biochem. 1998;67:199–225. - PubMed
-
- Brown M S, Goldstein J L. Cell. 1997;89:331–340. - PubMed
-
- Hua X, Wu J, Goldstein J L, Brown M S, Hobbs H H. Genomics. 1995;25:667–673. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

