Ligand-receptor binding measured by laser-scanning imaging
- PMID: 10500140
- PMCID: PMC17997
- DOI: 10.1073/pnas.96.20.11122
Ligand-receptor binding measured by laser-scanning imaging
Abstract
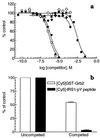
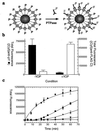
This report describes the integration of laser-scanning fluorometric cytometry and nonseparation ligand-binding techniques to provide new assay methods adaptable to miniaturization and high-throughput screening. Receptor-bound, cyanine dye-labeled ligands, [Cy]ligands, were discriminated from those free in solution by measuring the accumulated fluorescence associated with a receptor-containing particle. To illustrate the various binding formats accommodated by this technique, saturation- and competition-binding analyses were performed with [Cy]ligands and their cognate receptors expressed in CHO cells or as fusion proteins coated on polystyrene microspheres. We have successfully applied this technique to the analysis of G protein-coupled receptors, cytokine receptors, and SH2 domains. Multiparameter readouts from ligands labeled separately with Cy5 and Cy5.5 demonstrate the simultaneous analysis of two target receptors in a single well. In addition, laser-scanning cytometry has been used to assay enzymes such as phosphatases and in the development of single-step fluorescent immunoassays.
Figures



References
-
- Debouck C, Goodfellow P N. Nat Genet. 1999;21:48–50. - PubMed
-
- Chabala J C. In: Comb. Chem. Mol. Diversity Drug Discovery. Gordon E M, Kerwin J F Jr, editors. New York: Wiley–Liss; 1998. pp. 3–15.
-
- Hart H E, Greenwald E B. Mol Immunol. 1979;16:265–267. - PubMed
-
- Hemmila I, Webb S. Drug Discovery Today. 1997;2:373–381.
-
- Tsien R Y, Bacskai B J, Adams S R. Trends Cell Biol. 1993;3:242–245. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

