Halomethane:bisulfide/halide ion methyltransferase, an unusual corrinoid enzyme of environmental significance isolated from an aerobic methylotroph using chloromethane as the sole carbon source
- PMID: 10508052
- PMCID: PMC91570
- DOI: 10.1128/AEM.65.10.4301-4312.1999
Halomethane:bisulfide/halide ion methyltransferase, an unusual corrinoid enzyme of environmental significance isolated from an aerobic methylotroph using chloromethane as the sole carbon source
Abstract
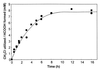
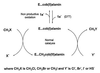
A novel dehalogenating/transhalogenating enzyme, halomethane:bisulfide/halide ion methyltransferase, has been isolated from the facultatively methylotrophic bacterium strain CC495, which uses chloromethane (CH(3)Cl) as the sole carbon source. Purification of the enzyme to homogeneity was achieved in high yield by anion-exchange chromatography and gel filtration. The methyltransferase was composed of a 67-kDa protein with a corrinoid-bound cobalt atom. The purified enzyme was inactive but was activated by preincubation with 5 mM dithiothreitol and 0.5 mM CH(3)Cl; then it catalyzed methyl transfer from CH(3)Cl, CH(3)Br, or CH(3)I to the following acceptor ions (in order of decreasing efficacy): I(-), HS(-), Cl(-), Br(-), NO(2)(-), CN(-), and SCN(-). Spectral analysis indicated that cobalt in the native enzyme existed as cob(II)alamin, which upon activation was reduced to the cob(I)alamin state and then was oxidized to methyl cob(III)alamin. During catalysis, the enzyme shuttles between the methyl cob(III)alamin and cob(I)alamin states, being alternately demethylated by the acceptor ion and remethylated by halomethane. Mechanistically the methyltransferase shows features in common with cobalamin-dependent methionine synthase from Escherichia coli. However, the failure of specific inhibitors of methionine synthase such as propyl iodide, N(2)O, and Hg(2+) to affect the methyltransferase suggests significant differences. During CH(3)Cl degradation by strain CC495, the physiological acceptor ion for the enzyme is probably HS(-), a hypothesis supported by the detection in cell extracts of methanethiol oxidase and formaldehyde dehydrogenase activities which provide a metabolic route to formate. 16S rRNA sequence analysis indicated that strain CC495 clusters with Rhizobium spp. in the alpha subdivision of the Proteobacteria and is closely related to strain IMB-1, a recently isolated CH(3)Br-degrading bacterium (T. L. Connell Hancock, A. M. Costello, M. E. Lidstrom, and R. S. Oremland, Appl. Environ. Microbiol. 64:2899-2905, 1998). The presence of this methyltransferase in bacterial populations in soil and sediments, if widespread, has important environmental implications.
Figures







References
-
- Andreae M O. Biomass burning: its history, use and distribution and its impact on environmental quality and global climate. In: Levine J S, editor. Global biomass burning. Cambridge, Mass: MIT Press; 1991. pp. 3–21.
-
- Anthony C. Assimilation of carbon by methylotrophs. In: Goldberg I, Rokem J S, editors. Biology of methylotrophs. Stoneham, Mass: Butterworth-Heinemann; 1991. pp. 79–109. - PubMed
-
- Attwood M M. Formaldehyde dehydrogenases from methylotrophs. Methods Enzymol. 1990;188:314–327.
-
- Banerjee R V, Harder S R, Ragsdale S W, Matthews R G. Mechanism of reductive activation of cobalamin-dependent methionine synthase: an electron paramagnetic resonance spectroelectrochemical study. Biochemistry. 1990;29:1129–1135. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

