Comparative analysis of noncoding regions of 77 orthologous mouse and human gene pairs
- PMID: 10508839
- PMCID: PMC310816
- DOI: 10.1101/gr.9.9.815
Comparative analysis of noncoding regions of 77 orthologous mouse and human gene pairs
Erratum in
- Genome Res 1999 Nov;9(11):1156
Abstract
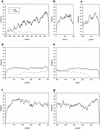
A data set of 77 genomic mouse/human gene pairs has been compiled from the EMBL nucleotide database, and their corresponding features determined. This set was used to analyze the degree of conservation of noncoding sequences between mouse and human. A new alignment algorithm was developed to cope with the fact that large parts of noncoding sequences are not alignable in a meaningful way because of genetic drift. This new algorithm, DNA Block Aligner (DBA), finds colinear-conserved blocks that are flanked by nonconserved sequences of varying lengths. The noncoding regions of the data set were aligned with DBA. The proportion of the noncoding regions covered by blocks >60% identical was 36% for upstream regions, 50% for 5' UTRs, 23% for introns, and 56% for 3' UTRs. These blocks of high identity were more or less evenly distributed across the length of the features, except for upstream regions in which the first 100 bp upstream of the transcription start site was covered in up to 70% of the gene pairs. This data set complements earlier sets on the basis of cDNA sequences and will be useful for further comparative studies. [This paper contains supplementary data that can be found at http://www.genome.org [corrected]].
Figures

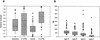
References
-
- Altschul S, Gish W, Miller W, Meyers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Ansari-Lari MA, Oeltjen JC, Schwartz S, Zhang Z, Muzny DM, Lu J, Gorrell JH, Chinault AC, Belmont JW, Miller W, Gibbs RA. Comparative sequence analysis of a gene-rich cluster at human chromosome 12p13 and its syntenic region in mouse chromosome 6. Genome Res. 1998;8:29–40. - PubMed
-
- Bernardi G. The isochore organization of the human genome and its evolutionary history–A review. Gene. 1993;135:57–66. - PubMed
-
- Birney E, Durbin R. Dynamite: A flexible code generating language for dynamic programming methods used in sequence comparison. Intell Syst Mol Biol. 1997;5:56–64. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
