Spatial relationship between transcription sites and chromosome territories
- PMID: 10508851
- PMCID: PMC2164981
- DOI: 10.1083/jcb.147.1.13
Spatial relationship between transcription sites and chromosome territories
Abstract
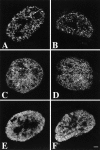
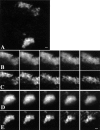
We have investigated the spatial relationship between transcription sites and chromosome territories in the interphase nucleus of human female fibroblasts. Immunolabeling of nascent RNA was combined with visualization of chromosome territories by fluorescent in situ hybridization (FISH). Transcription sites were found scattered throughout the territory of one of the two X chromosomes, most likely the active X chromosome, and that of both territories of chromosome 19. The other X chromosome territory, probably the inactive X chromosome, was devoid of transcription sites. A distinct substructure was observed in interphase chromosome territories. Intensely labeled subchromosomal domains are surrounded by less strongly labeled areas. The intensely labeled domains had a diameter in the range of 300-450 nm and were sometimes interconnected, forming thread-like structures. Similar large scale chromatin structures were observed in HeLa cells expressing green fluorescent protein (GFP)-tagged histone H2B. Strikingly, nascent RNA was almost exclusively found in the interchromatin areas in chromosome territories and in between strongly GFP-labeled chromatin domains. These observations support a model in which transcriptionally active chromatin in chromosome territories is markedly compartmentalized. Active loci are located predominantly at or near the surface of compact chromatin domains, depositing newly synthesized RNA directly into the interchromatin space.
Figures



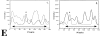


References
-
- Belmont A.S., Braunfield M.B., Sedat J.W., Agard D.A. Large-scale Chromatin structural domains within mitotic and interphase chromosomes in vivo and in vitro. Chromosoma. 1989;98:129–143. - PubMed
-
- Brakenhoff G.J., Van der Voort H.T.M., Van Spronsen E.A., Linnemans W.A.M., Nanninga N. Three-dimensional chromatin distribution in neuroblastoma nuclei shown by confocal scanning laser microscopy. Nature. 1985;317:748–749. - PubMed
-
- Braunstein M., Rose A.B., Holmes S.G., Allis C.D., Broach J.R. Transcriptional silencing in yeast is associated with reduced histone acetylation. Genes Dev. 1993;7:592–604. - PubMed

