Antifungal activities of antineoplastic agents: Saccharomyces cerevisiae as a model system to study drug action
- PMID: 10515904
- PMCID: PMC88926
- DOI: 10.1128/CMR.12.4.583
Antifungal activities of antineoplastic agents: Saccharomyces cerevisiae as a model system to study drug action
Abstract
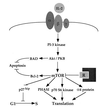
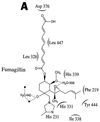
Recent evolutionary studies reveal that microorganisms including yeasts and fungi are more closely related to mammals than was previously appreciated. Possibly as a consequence, many natural-product toxins that have antimicrobial activity are also toxic to mammalian cells. While this makes it difficult to discover antifungal agents without toxic side effects, it also has enabled detailed studies of drug action in simple genetic model systems. We review here studies on the antifungal actions of antineoplasmic agents. Topics covered include the mechanisms of action of inhibitors of topoisomerases I and II; the immunosuppressants rapamycin, cyclosporin A, and FK506; the phosphatidylinositol 3-kinase inhibitor wortmannin; the angiogenesis inhibitors fumagillin and ovalicin; the HSP90 inhibitor geldanamycin; and agents that inhibit sphingolipid metabolism. In general, these natural products inhibit target proteins conserved from microorganisms to humans. These studies highlight the potential of microorganisms as screening tools to elucidate the mechanisms of action of novel pharmacological agents with unique effects against specific mammalian cell types, including neoplastic cells. In addition, this analysis suggests that antineoplastic agents and derivatives might find novel indications in the treatment of fungal infections, for which few agents are presently available, toxicity remains a serious concern, and drug resistance is emerging.
Figures


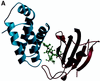
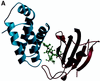



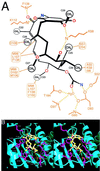

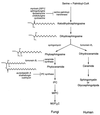
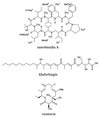

Similar articles
-
Signal-transduction cascades as targets for therapeutic intervention by natural products.Trends Biotechnol. 1998 Oct;16(10):427-33. doi: 10.1016/s0167-7799(98)01239-6. Trends Biotechnol. 1998. PMID: 9807840 Review.
-
Synergistic antifungal activities of bafilomycin A(1), fluconazole, and the pneumocandin MK-0991/caspofungin acetate (L-743,873) with calcineurin inhibitors FK506 and L-685,818 against Cryptococcus neoformans.Antimicrob Agents Chemother. 2000 Mar;44(3):739-46. doi: 10.1128/AAC.44.3.739-746.2000. Antimicrob Agents Chemother. 2000. PMID: 10681348 Free PMC article.
-
Teaching old drugs new tricks: reincarnating immunosuppressants as antifungal drugs.Curr Opin Investig Drugs. 2003 Feb;4(2):192-9. Curr Opin Investig Drugs. 2003. PMID: 12669381 Review.
-
Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans.Mol Cell Biol. 1999 Jun;19(6):4101-12. doi: 10.1128/MCB.19.6.4101. Mol Cell Biol. 1999. PMID: 10330150 Free PMC article.
-
Protein kinase activity and identification of a toxic effector domain of the target of rapamycin TOR proteins in yeast.Mol Biol Cell. 1999 Aug;10(8):2531-46. doi: 10.1091/mbc.10.8.2531. Mol Biol Cell. 1999. PMID: 10436010 Free PMC article.
Cited by
-
Ibuprofen potentiates the in vivo antifungal activity of fluconazole against Candida albicans murine infection.Antimicrob Agents Chemother. 2015 Jul;59(7):4289-92. doi: 10.1128/AAC.05056-14. Epub 2015 Apr 6. Antimicrob Agents Chemother. 2015. PMID: 25845879 Free PMC article.
-
The genome of tolypocladium inflatum: evolution, organization, and expression of the cyclosporin biosynthetic gene cluster.PLoS Genet. 2013 Jun;9(6):e1003496. doi: 10.1371/journal.pgen.1003496. Epub 2013 Jun 20. PLoS Genet. 2013. PMID: 23818858 Free PMC article.
-
Direct effects of non-antifungal agents used in cancer chemotherapy and organ transplantation on the development and virulence of Candida and Aspergillus species.Virulence. 2011 Jul-Aug;2(4):280-95. doi: 10.4161/viru.2.4.16764. Epub 2011 Jul 1. Virulence. 2011. PMID: 21701255 Free PMC article. Review.
-
Target of rapamycin (TOR) in nutrient signaling and growth control.Genetics. 2011 Dec;189(4):1177-201. doi: 10.1534/genetics.111.133363. Genetics. 2011. PMID: 22174183 Free PMC article. Review.
-
Antifungal mechanism of volatile compounds emitted by Actinomycetota Paenarthrobacter ureafaciens from a disease-suppressive soil on Saccharomyces cerevisiae.mSphere. 2023 Oct 24;8(5):e0032423. doi: 10.1128/msphere.00324-23. Epub 2023 Sep 26. mSphere. 2023. PMID: 37750721 Free PMC article.
References
-
- Abe J, Zhou W, Takuwa N, Taguchi J, Kurokawa K, Kumada M, Takuwa Y. A fumagillin derivative angiogenesis inhibitor, AGM-1470, inhibits activation of cyclin-dependent kinases and phosphorylation of retinoblastoma gene product but not protein tyrosyl phosphorylation or protooncogene expression in vascular endothelial cells. Cancer Res. 1994;54:3407–3412. - PubMed
-
- Aitken A, Klee C B, Cohen P. The structure of the B subunit of calcineurin. Eur J Biochem. 1984;139:663–671. - PubMed
-
- Alarcon C M, Cardenas M E, Heitman J. Mammalian RAFT1 kinase domain provides rapamycin-sensitive TOR function in yeast. Genes Dev. 1996;10:279–288. - PubMed
-
- Alexander B D, Perfect J R. Antifungal resistance trends towards the year 2000. Implications for therapy and new approaches. Drugs. 1997;54:657–678. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

