Regulation of the transposase of Tn4652 by the transposon-encoded protein TnpC
- PMID: 10515920
- PMCID: PMC103765
- DOI: 10.1128/JB.181.20.6312-6318.1999
Regulation of the transposase of Tn4652 by the transposon-encoded protein TnpC
Abstract
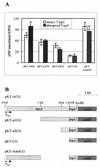
Transposition is a DNA reorganization reaction potentially deleterious for the host. The frequency of transposition is limited by the amount of transposase. Therefore, strict regulation of a transposase is required to keep control over the destructive multiplication of the mobile element. We have shown previously that the expression of the transposase (tnpA) of the Pseudomonas putida PaW85 transposon Tn4652 is positively affected by integration host factor. Here, we present evidence that the amount of the transposase of Tn4652 in P. putida cells is controlled by the transposon-encoded protein (TnpC). Sequence analysis of the 120-amino-acid-long TnpC, coded just downstream of the tnpA gene, showed that it has remarkable similarity to the putative polypeptide encoded by the mercury resistance transposon Tn5041. As determined by quantitative Western blot analysis, the abundance of TnpA was reduced up to 10-fold in the intact tnpC background. In vivo experiments using transcriptional and translational fusions of the tnpA gene and the reporter gene gusA indicated that TnpC operates in the regulation of the transposase of Tn4652 at the post-transcriptional level.
Figures


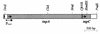


References
-
- Allison R G, Chaconas G. Role of the A protein-binding sites in the in vitro transposition of Mu DNA. J Biol Chem. 1992;267:19963–19970. - PubMed
-
- Arini A, Keller M P, Arber W. An antisense RNA in IS30 regulates the translational expression of the transposase. Biol Chem. 1997;378:1421–1431. - PubMed
-
- Bagdasarian M M, Amann E, Lurz R, Ruckert B, Bagdasarian M. Activity of the hybrid trp-lac(tac) promoter of Escherichia coli in Pseudomonas putida. Construction of broad-host-range, controlled-expression vectors. Gene. 1983;26:273–282. - PubMed
-
- Bayley S A, Duggleby C J, Worsey M J, Williams P A, Hardy K G, Broda P. Two modes of loss of the TOL function from Pseudomonas putida mt-2. Mol Gen Genet. 1977;154:203–204. - PubMed
-
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

