Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid
- PMID: 10515926
- PMCID: PMC103771
- DOI: 10.1128/JB.181.20.6361-6370.1999
Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid
Abstract
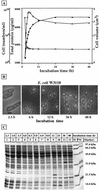
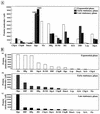
The genome DNA of Escherichia coli is associated with about 10 DNA-binding structural proteins, altogether forming the nucleoid. The nucleoid proteins play some functional roles, besides their structural roles, in the global regulation of such essential DNA functions as replication, recombination, and transcription. Using a quantitative Western blot method, we have performed for the first time a systematic determination of the intracellular concentrations of 12 species of the nucleoid protein in E. coli W3110, including CbpA (curved DNA-binding protein A), CbpB (curved DNA-binding protein B, also known as Rob [right origin binding protein]), DnaA (DNA-binding protein A), Dps (DNA-binding protein from starved cells), Fis (factor for inversion stimulation), Hfq (host factor for phage Q(beta)), H-NS (histone-like nucleoid structuring protein), HU (heat-unstable nucleoid protein), IciA (inhibitor of chromosome initiation A), IHF (integration host factor), Lrp (leucine-responsive regulatory protein), and StpA (suppressor of td mutant phenotype A). Intracellular protein levels reach a maximum at the growing phase for nine proteins, CbpB (Rob), DnaA, Fis, Hfq, H-NS, HU, IciA, Lrp, and StpA, which may play regulatory roles in DNA replication and/or transcription of the growth-related genes. In descending order, the level of accumulation, calculated in monomers, in growing E. coli cells is Fis, Hfq, HU, StpA, H-NS, IHF*, CbpB (Rob), Dps*, Lrp, DnaA, IciA, and CbpA* (stars represent the stationary-phase proteins). The order of abundance, in descending order, in the early stationary phase is Dps*, IHF*, HU, Hfq, H-NS, StpA, CbpB (Rob), DnaA, Lrp, IciA, CbpA, and Fis, while that in the late stationary phase is Dps*, IHF*, Hfq, HU, CbpA*, StpA, H-NS, CbpB (Rob), DnaA, Lrp, IciA, and Fis. Thus, the major protein components of the nucleoid change from Fis and HU in the growing phase to Dps in the stationary phase. The curved DNA-binding protein, CbpA, appears only in the late stationary phase. These changes in the composition of nucleoid-associated proteins in the stationary phase are accompanied by compaction of the genome DNA and silencing of the genome functions.
Figures

References
-
- Almirón M, Link A J, Furlong D, Kolter R. A novel DNA binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. - PubMed
-
- Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. - PubMed
-
- Craig N L, Nash S E. E. coli integration host factor binds to specific sites in DNA. Cell. 1984;39:707–716. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

