A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus
- PMID: 10515939
- PMCID: PMC103784
- DOI: 10.1128/JB.181.20.6469-6477.1999
A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus
Abstract
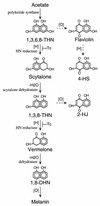
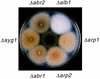
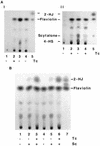
Aspergillus fumigatus, a filamentous fungus producing bluish-green conidia, is an important opportunistic pathogen that primarily affects immunocompromised patients. Conidial pigmentation of A. fumigatus significantly influences its virulence in a murine model. In the present study, six genes, forming a gene cluster spanning 19 kb, were identified as involved in conidial pigment biosynthesis in A. fumigatus. Northern blot analyses showed the six genes to be developmentally regulated and expressed during conidiation. The gene products of alb1 (for "albino 1"), arp1 (for "aspergillus reddish-pink 1"), and arp2 have high similarity to polyketide synthases, scytalone dehydratases, and hydroxynaphthalene reductases, respectively, found in the dihydroxynaphthalene (DHN)-melanin pathway of brown and black fungi. The abr1 gene (for "aspergillus brown 1") encodes a putative protein possessing two signatures of multicopper oxidases. The abr2 gene product has homology to the laccase encoded by the yA gene of Aspergillus nidulans. The function of ayg1 (for "aspergillus yellowish-green 1") remains unknown. Involvement of the six genes in conidial pigmentation was confirmed by the altered conidial color phenotypes that resulted from disruption of each gene in A. fumigatus. The presence of a DHN-melanin pathway in A. fumigatus was supported by the accumulation of scytalone and flaviolin in the arp1 deletant, whereas only flaviolin was accumulated in the arp2 deletants. Scytalone and flaviolin are well-known signature metabolites of the DHN-melanin pathway. Based on DNA sequence similarity, gene disruption results, and biochemical analyses, we conclude that the 19-kb DNA fragment contains a six-gene cluster which is required for conidial pigment biosynthesis in A. fumigatus. However, the presence of abr1, abr2, and ayg1 in addition to alb1, arp1, and arp2 suggests that conidial pigment biosynthesis in A. fumigatus is more complex than the known DHN-melanin pathway.
Figures





References
-
- Amaar Y G, Moore M M. Mapping of the nitrate-assimilation gene cluster (crnA-niiA-niaD) and characterization of the nitrite reductase gene (niiA) in the opportunistic fungal pathogen Aspergillus fumigatus. Curr Genet. 1998;33:206–215. - PubMed
-
- Bell A A, Stipanovic R D, Puhalla J E. Pentaketide metabolites of Verticillium dahliae: identification of (+)-scytalone as a natural precursor to melanin. Tetrahedron. 1976;32:1353–1356.
-
- Bell A A, Wheeler M H. Biosynthesis and functions of fungal melanins. Annu Rev Phytopathol. 1986;24:411–451.
-
- Bennett J W, Papa K E. The aflatoxigenic Aspergillus species. Adv Plant Pathol. 1988;6:263–280.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

