An Mpsi-containing heterologous RNA, but not env mRNA, is efficiently packaged into avian retroviral particles
- PMID: 10515997
- PMCID: PMC112923
- DOI: 10.1128/JVI.73.11.8926-8933.1999
An Mpsi-containing heterologous RNA, but not env mRNA, is efficiently packaged into avian retroviral particles
Abstract
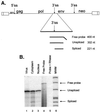
Retroviruses preferentially package full-length genomic RNA over spliced viral messages. For most retroviruses, this preference is likely due to the absence of all or part of the packaging signal on subgenomic RNAs. In avian leukosis-sarcoma virus, however, we have shown that the minimal packaging signal, MPsi, is located upstream of the 5' splice site and therefore is present on both genomic and spliced RNAs. We now show that an MPsi-containing heterologous RNA is packaged only 2.6-fold less efficiently than genomic Rous sarcoma virus RNA. Thus, few additional packaging sequences and/or structures exist outside of MPsi. In contrast, we found that env mRNA is not efficiently packaged. These results indicate that either MPsi is not functional on this RNA or the RNA is somehow segregated from the packaging machinery. Finally, deletion of sequences from the 3' end of MPsi was found to reduce the packaging efficiency of heterologous RNAs.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

