Nonreplicative RNA recombination in poliovirus
- PMID: 10516001
- PMCID: PMC112927
- DOI: 10.1128/JVI.73.11.8958-8965.1999
Nonreplicative RNA recombination in poliovirus
Abstract
Current models of recombination between viral RNAs are based on replicative template-switch mechanisms. The existence of nonreplicative RNA recombination in poliovirus is demonstrated in the present study by the rescue of viable viruses after cotransfections with different pairs of genomic RNA fragments with suppressed translatable and replicating capacities. Approximately 100 distinct recombinant genomes have been identified. The majority of crossovers occurred between nonhomologous segments of the partners and might have resulted from transesterification reactions, not necessarily involving an enzymatic activity. Some of the crossover loci are clustered. The origin of some of these "hot spots" could be explained by invoking structures similar to known ribozymes. A significant proportion of recombinant RNAs contained the entire 5' partner, if its 3' end was oxidized or phosphorylated prior to being mixed with the 3' partner. All of these observations are consistent with a mechanism that involves intermediary formation of the 2',3'-cyclic phosphate and 5'-hydroxyl termini. It is proposed that nonreplicative RNA recombination may contribute to evolutionarily significant RNA rearrangements.
Figures
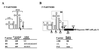


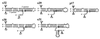
References
-
- Adams R L P, Knowler J T, Leader D P. The biochemistry of the nucleic acids. 10th ed. London, England: Chapman and Hall; 1986.
-
- Agol V I. Recombination and other genomic rearrangements in picornaviruses. Semin Virol. 1997;8:1–9.
-
- Andino R, Rieckhof G E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. - PubMed
-
- Birikh K R, Heaton P A, Eckstein F. The structure, function and application of the hammerhead ribozyme. Eur J Biochem. 1997;245:1–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

