Poliovirus mutants at histidine 195 of VP2 do not cleave VP0 into VP2 and VP4
- PMID: 10516013
- PMCID: PMC112939
- DOI: 10.1128/JVI.73.11.9072-9079.1999
Poliovirus mutants at histidine 195 of VP2 do not cleave VP0 into VP2 and VP4
Abstract
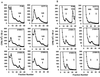
The final stage of poliovirus assembly is characterized by a cleavage of the capsid precursor protein VP0 into VP2 and VP4. This cleavage is thought to be autocatalytic and dependent on RNA encapsidation. Analysis of the poliovirus empty capsid structure has led to a mechanistic model for VP0 cleavage involving a conserved histidine residue that is present in the surrounding environment of the VP0 cleavage site. Histidine 195 of VP2 (2195H) is hypothesized to activate local water molecules, thus initiating a nucleophilic attack at the scissile bond. To test this hypothesis, 2195H mutants were constructed and their phenotypes were characterized. Consistent with the requirement of VP0 cleavage for poliovirus infectivity, all 2195H mutants were nonviable upon introduction of the mutant genomes into HeLa cells. Replacement of 2195H with threonine or arginine resulted in the assembly of a highly unstable 150S virus particle. Further analyses showed that these particles contain genomic RNA and uncleaved VP0, criteria associated with the provirion assembly intermediate. These data support the involvement of 2195H in mediating VP0 cleavage during the final stages of virus assembly.
Figures
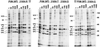




Similar articles
-
Amino acid substitutions in the poliovirus maturation cleavage site affect assembly and result in accumulation of provirions.J Virol. 1995 Mar;69(3):1540-7. doi: 10.1128/JVI.69.3.1540-1547.1995. J Virol. 1995. PMID: 7853487 Free PMC article.
-
Myristylation of poliovirus capsid precursor P1 is required for assembly of subviral particles.J Virol. 1992 Jul;66(7):4556-63. doi: 10.1128/JVI.66.7.4556-4563.1992. J Virol. 1992. PMID: 1318418 Free PMC article.
-
Catalysis of poliovirus VP0 maturation cleavage is not mediated by serine 10 of VP2.J Virol. 1991 Jan;65(1):326-34. doi: 10.1128/JVI.65.1.326-334.1991. J Virol. 1991. PMID: 1845893 Free PMC article.
-
Mutations in the poliovirus P1 capsid precursor at arginine residues VP4-ARG34, VP3-ARG223, and VP1-ARG129 affect virus assembly and encapsidation of genomic RNA.Virology. 1994 Feb 15;199(1):20-34. doi: 10.1006/viro.1994.1094. Virology. 1994. PMID: 8116243
-
Hepatitis A Virus Capsid Structure.Cold Spring Harb Perspect Med. 2019 May 1;9(5):a031807. doi: 10.1101/cshperspect.a031807. Cold Spring Harb Perspect Med. 2019. PMID: 30037986 Free PMC article. Review.
Cited by
-
Mechanism of enterovirus VP0 maturation cleavage based on the structure of a stabilised assembly intermediate.PLoS Pathog. 2024 Sep 19;20(9):e1012511. doi: 10.1371/journal.ppat.1012511. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39298524 Free PMC article.
-
Strategy for nonenveloped virus entry: a hydrophobic conformer of the reovirus membrane penetration protein micro 1 mediates membrane disruption.J Virol. 2002 Oct;76(19):9920-33. doi: 10.1128/jvi.76.19.9920-9933.2002. J Virol. 2002. PMID: 12208969 Free PMC article.
-
Enterovirus-like particles encapsidate RNA and exhibit decreased stability due to lack of maturation.PLoS Pathog. 2025 Feb 4;21(2):e1012873. doi: 10.1371/journal.ppat.1012873. eCollection 2025 Feb. PLoS Pathog. 2025. PMID: 39903789 Free PMC article.
-
Cellular N-myristoyltransferases play a crucial picornavirus genus-specific role in viral assembly, virion maturation, and infectivity.PLoS Pathog. 2018 Aug 6;14(8):e1007203. doi: 10.1371/journal.ppat.1007203. eCollection 2018 Aug. PLoS Pathog. 2018. PMID: 30080883 Free PMC article.
-
Vaccine Potency and Structure of Yeast-Produced Polio Type 2 Stabilized Virus-like Particles.Vaccines (Basel). 2024 Sep 20;12(9):1077. doi: 10.3390/vaccines12091077. Vaccines (Basel). 2024. PMID: 39340107 Free PMC article.
References
-
- Acharya R, Fry E, Stuart D, Fox G, Rowlands D, Brown F. The three-dimensional structure of foot-and-mouth disease virus at 2.9Å resolution. Nature. 1989;337:709–716. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

