Site-directed mutagenesis of the virion host shutoff gene (UL41) of herpes simplex virus (HSV): analysis of functional differences between HSV type 1 (HSV-1) and HSV-2 alleles
- PMID: 10516018
- PMCID: PMC112944
- DOI: 10.1128/JVI.73.11.9117-9129.1999
Site-directed mutagenesis of the virion host shutoff gene (UL41) of herpes simplex virus (HSV): analysis of functional differences between HSV type 1 (HSV-1) and HSV-2 alleles
Abstract
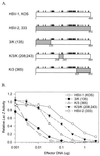
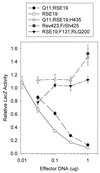
During lytic herpes simplex virus (HSV) infections, the HSV virion host shutoff protein (UL41) accelerates the turnover of host and viral mRNAs. Although the UL41 polypeptides from HSV type 1 (HSV-1) strain KOS and HSV-2 strain 333 are 87% identical, HSV-2 strains generally shut off the host more rapidly and completely than HSV-1 strains. In a previous study, we identified three regions of the HSV-2 UL41 polypeptide (amino acids 1 to 135, 208 to 243, and 365 to 492) that enhance the activity of KOS when substituted for the corresponding portions of the KOS protein (D. N. Everly, Jr., and G. S. Read, J. Virol. 71:7157-7166, 1997). These results have been extended through the analysis of more than 50 site-directed mutants of UL41 in which selected HSV-2 amino acids were introduced into an HSV-1 background and HSV-1 amino acids were introduced into the HSV-2 allele. The HSV-2 amino acids R22 and E25 were found to contribute dramatically to the greater activity of the HSV-2 allele, as did the HSV-2 amino acids A396 and S423. The substitution of six HSV-2 amino acids between residues 210 and 242 enhanced the HSV-1 activity to a lesser extent. In most cases, individual substitutions or the substitution of combinations of fewer than all six amino acids reduced the UL41 activity to less than that of KOS. The results pinpoint several type-specific amino acids that are largely responsible for the greater activity of the UL41 polypeptide of HSV-2. In addition, several spontaneous mutations that abolish detectable UL41 activity were identified.
Figures








References
-
- Becker Y, Tavor E, Asher Y, Berkowiltz C, Moyal M. Effect of herpes simplex virus type-1 UL41 gene on the stability of mRNA from the cellular genes: beta-actin, fibronectin, glucose transporter-1, and docking protein, and on virus intraperitoneal pathogenicity of newborn mice. Virus Genes. 1993;7:133–143. - PubMed
-
- Berthomme H, Jacquemont B, Epstein A. The pseudorabies virus host-shutoff homolog gene: nucleotide sequence and comparison with alphaherpesvirus protein counterparts. Virology. 1993;193:1028–1032. - PubMed
-
- Darteil R, Bublot M, Laplace E, Bouquet J F, Audonnet J C, Riviere M. Herpesvirus of turkey recombinant viruses expressing infectious bursal disease virus (IBDV) VP2 immunogen induce protection against an IBDV virulent challenge in chickens. Virology. 1995;211:481–490. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

