Replication-defective bovine adenovirus type 3 as an expression vector
- PMID: 10516020
- PMCID: PMC112946
- DOI: 10.1128/JVI.73.11.9137-9144.1999
Replication-defective bovine adenovirus type 3 as an expression vector
Abstract
Although recombinant human adenovirus (HAV)-based vectors offer several advantages for somatic gene therapy and vaccination over other viral vectors, it would be desirable to develop alternative vectors with prolonged expression and decreased toxicity. Toward this objective, a replication-defective bovine adenovirus type 3 (BAV-3) was developed as an expression vector. Bovine cell lines designated VIDO R2 (HAV-5 E1A/B-transformed fetal bovine retina cell [FBRC] line) and 6.93.9 (Madin-Darby bovine kidney [MDBK] cell line expressing E1 proteins) were developed and found to complement the E1A deletion in BAV-3. Replication-defective BAV-3 with a 1.7-kb deletion removing most of the E1A and E3 regions was constructed. This virus could be grown in VIDO R2 or 6.93.9 cells but not in FBRC or MDBK cells. The results demonstrated that the E1 region of HAV-5 has the capacity to transform bovine retina cells and that the E1A region of HAV-5 can complement that of BAV-3. A replication-defective BAV-3 vector expressing bovine herpesvirus type 1 glycoprotein D from the E1A region was made. A similar replication-defective vector expressing the hemagglutinin-esterase gene of bovine coronavirus from the E3 region was isolated. Although these viruses grew less efficiently than the replication-competent recombinant BAV-3 (E3 deleted), they are suitable for detailed studies with animals to evaluate the safety, duration of foreign gene expression, and ability to induce immune responses. In addition, a replication-competent recombinant BAV-3 expressing green fluorescent protein was constructed and used to evaluate the host range of BAV-3 under cell culture conditions. The development of bovine E1A-complementing cell lines and the generation of replication-defective BAV-3 vectors is a major technical advancement for defining the use of BAV-3 as vector for vaccination against diseases of cattle and somatic gene therapy in humans.
Figures



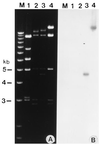
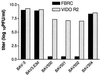
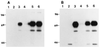

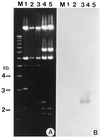
References
-
- Bartha A. Proposal for subgrouping of bovine adenoviruses. Acta Vet. 1969;19:319–321. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

