Nonrandom distribution of hepatitis C virus quasispecies in plasma and peripheral blood mononuclear cell subsets
- PMID: 10516029
- PMCID: PMC112955
- DOI: 10.1128/JVI.73.11.9213-9221.1999
Nonrandom distribution of hepatitis C virus quasispecies in plasma and peripheral blood mononuclear cell subsets
Abstract
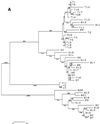
The existence of an extrahepatic reservoir of hepatitis C virus (HCV) is suggested by differences in quasispecies composition between the liver, peripheral blood mononuclear cells, and serum. We studied HCV RNA compartmentalization in the plasma of nine patients, in CD19(+), CD8(+), and CD4(+) positively selected cells, and also in the negatively selected cell fraction (NF). HCV RNA was detected in all plasma samples, in seven of nine CD19(+), three of eight CD8(+), and one of nine CD4(+) cell samples, and in seven of eight NF cells. Cloning and sequencing of HVR1 in two patients showed a sequence grouping: quasispecies from a given compartment (all studied compartments for one patient and CD8(+) and NF for the other) were statistically more genetically like each other than like quasispecies from any other compartment. The characteristics of amino acid and nucleotide substitutions suggested the same structural constraints on HVR1, even in very divergent strains from the cellular compartments, and homogeneous selection pressure on the different compartments. These findings demonstrate the compartmental distribution of HCV quasispecies within peripheral blood cell subsets and have important implications for the study of extrahepatic HCV replication and interaction with the immune system.
Figures


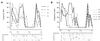
References
-
- Barnaba V, Franco A, Alberti A, Benvenuto R, Balsano F. Selective killing of hepatitis B envelope antigen-specific B cells by class I-restricted, exogenous antigen-specific T lymphocytes. Nature. 1990;345:258–260. - PubMed
-
- Chen M, Sällberg M, Sönnerborg A, Weiland O, Mattsson L, Jin L, Birkett A, Peterson D, Milich D R. Limited humoral immunity in hepatitis C virus infection. Gastroenterology. 1999;116:135–143. - PubMed
-
- Choo Q L, Kuo G, Weiner A J, Overby L R, Bradly D W, Houghton M. Isolation of a cDNA clone derived from blood-borne non-A non-B viral hepatitis genome. Science. 1989;244:359–362. - PubMed
-
- Choo S H, So H S, Cho J M, Ryu W S. Association of hepatitis C virus particles with immunoglobulin: a mechanism for persistent infection. J Gen Virol. 1995;76:2337–2341. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

