Picornavirus internal ribosome entry site elements target RNA cleavage events induced by the herpes simplex virus virion host shutoff protein
- PMID: 10516030
- PMCID: PMC112956
- DOI: 10.1128/JVI.73.11.9222-9231.1999
Picornavirus internal ribosome entry site elements target RNA cleavage events induced by the herpes simplex virus virion host shutoff protein
Abstract
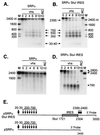
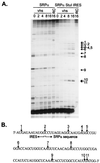
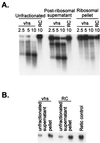
The herpes simplex virus (HSV) virion host shutoff (vhs) protein (UL41 gene product) is a component of the HSV virion tegument that triggers shutoff of host protein synthesis and accelerated mRNA degradation during the early stages of HSV infection. vhs displays weak amino acid sequence similarity to the fen-1 family of nucleases and suffices to induce accelerated RNA turnover through endoribonucleolytic cleavage events when it is expressed as the only HSV protein in a rabbit reticulocyte in vitro translation system. Although vhs selectively targets mRNAs in vivo, the basis for this selectivity remains obscure, since in vitro activity is not influenced by the presence of a 5' cap or 3' poly(A) tail. Here we show that vhs activity is greatly altered by placing an internal ribosome entry site (IRES) from encephalomyocarditis virus or poliovirus in the RNA substrate. Transcripts bearing the IRES were preferentially cleaved by the vhs-dependent endoribonuclease at multiple sites clustered in a narrow zone located immediately downstream of the element in a reaction that did not require ribosomes. Targeting was observed when the IRES was located at the 5' end or placed at internal sites in the substrate, indicating that it is independent of position or sequence context. These data indicate that the vhs-dependent nuclease can be selectively targeted by specific cis-acting elements in the RNA substrate, possibly through secondary structure or a component of the translational machinery.
Figures





References
-
- Buckley B, Ehrenfeld E. The cap-binding protein complex in uninfected and poliovirus-infected HeLa cells. J Biol Chem. 1987;262:13599–13606. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

