Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells
- PMID: 10516043
- PMCID: PMC112969
- DOI: 10.1128/JVI.73.11.9348-9361.1999
Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells
Abstract
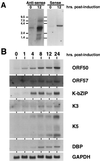
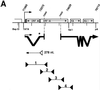
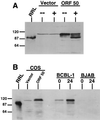
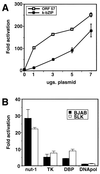
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) is a lymphotropic virus strongly linked to the development of KS, an endothelial cell neoplasm frequent in persons with AIDS. Reactivation from latency in B cells is thought to be an important antecedent to viral spread to endothelial cells during KS pathogenesis. Earlier experiments have posited a role for the transcriptional activator encoded by KSHV open reading frame 50 (ORF50) in such reactivation, since ectopic overexpression of this protein induces reactivation in latently infected B cells. Here we have explored several aspects of the expression, structure, and function of this protein bearing on this role. The ORF50 gene is expressed very early in lytic reactivation, before several other genes implicated as candidate regulatory genes in related viruses, and its expression can upregulate their promoters in transient assays. The protein is extensively phosphorylated in vivo and bears numerous sites for phosphorylation by protein kinase C, activators of which are potent stimulators of lytic induction. The C terminus of the ORF50 protein contains a domain that can strongly activate transcription when targeted to DNA; deletion of this domain generates an allele that expresses a truncated protein which retains the ability to form multimers with full-length ORF50 and functions as a dominant-negative protein. Expression of this allele in latently infected cells ablates spontaneous reactivation from latency and strikingly suppresses viral replication induced by multiple stimuli, including phorbol ester, ionomycin, and sodium butyrate. These results indicate that the ORF50 gene product plays an essential role in KSHV lytic replication and are consistent with its action as a putative molecular switch controlling the induction of virus from latency.
Figures



Similar articles
-
CCAAT/enhancer-binding protein-alpha is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters.J Virol. 2003 Sep;77(17):9590-612. doi: 10.1128/jvi.77.17.9590-9612.2003. J Virol. 2003. PMID: 12915572 Free PMC article.
-
Identification of Novel Kaposi's Sarcoma-Associated Herpesvirus Orf50 Transcripts: Discovery of New RTA Isoforms with Variable Transactivation Potential.J Virol. 2016 Dec 16;91(1):e01434-16. doi: 10.1128/JVI.01434-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795414 Free PMC article.
-
X box binding protein XBP-1s transactivates the Kaposi's sarcoma-associated herpesvirus (KSHV) ORF50 promoter, linking plasma cell differentiation to KSHV reactivation from latency.J Virol. 2007 Dec;81(24):13578-86. doi: 10.1128/JVI.01663-07. Epub 2007 Oct 10. J Virol. 2007. PMID: 17928342 Free PMC article.
-
The Rta/Orf50 transactivator proteins of the gamma-herpesviridae.Curr Top Microbiol Immunol. 2007;312:71-100. doi: 10.1007/978-3-540-34344-8_3. Curr Top Microbiol Immunol. 2007. PMID: 17089794 Review.
-
Lytic cycle switches of oncogenic human gammaherpesviruses.Adv Cancer Res. 2007;97:81-109. doi: 10.1016/S0065-230X(06)97004-3. Adv Cancer Res. 2007. PMID: 17419942 Review.
Cited by
-
Linear ubiquitination regulates the KSHV replication and transcription activator protein to control infection.Nat Commun. 2024 Jun 29;15(1):5515. doi: 10.1038/s41467-024-49887-6. Nat Commun. 2024. PMID: 38951495 Free PMC article.
-
Identification and characterization of the Orf49 protein of Kaposi's sarcoma-associated herpesvirus.J Virol. 2006 Mar;80(6):3062-70. doi: 10.1128/JVI.80.6.3062-3070.2006. J Virol. 2006. PMID: 16501115 Free PMC article.
-
Disruption of gammaherpesvirus 68 gene 50 demonstrates that Rta is essential for virus replication.J Virol. 2003 May;77(10):5731-9. doi: 10.1128/jvi.77.10.5731-5739.2003. J Virol. 2003. PMID: 12719566 Free PMC article.
-
beta-Adrenoreceptors reactivate Kaposi's sarcoma-associated herpesvirus lytic replication via PKA-dependent control of viral RTA.J Virol. 2005 Nov;79(21):13538-47. doi: 10.1128/JVI.79.21.13538-13547.2005. J Virol. 2005. PMID: 16227274 Free PMC article.
-
Systematic identification of cellular signals reactivating Kaposi sarcoma-associated herpesvirus.PLoS Pathog. 2007 Mar;3(3):e44. doi: 10.1371/journal.ppat.0030044. PLoS Pathog. 2007. PMID: 17397260 Free PMC article.
References
-
- Ambroziak J, Blackbourn D, Herndier B, Glogan R, Gullet J, McDonald A, Lennette E, Levy J. Herpesvirus-like sequences in HIV-infected and uninfected Kaposi’s sarcoma patients. Science. 1995;268:582–583. - PubMed
-
- Arvanitakis L, Geras-Raaka E, Varma A, Gershengorn M C, Cesarman E. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature. 1997;385:347–350. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1997.
-
- Bais C, Santomasso B, Coso O, Arvanitakis L, Raaka E G, Gutkind J S, Asch A S, Cesarman E, Gershengorn M C, Mesri E A. G-protein-coupled receptor of Kaposi’s sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature. 1998;391:86–89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

